Triton resource helps 'track' how kinesin molecules move
Researchers at UC San Diego’s Department of Chemistry and Biochemistry, in collaboration with several universities in the U.S., United Kingdom, and Poland, have developed a new picture of how kinesin molecules move along microtubules, or tiny biological train tracks – and how they sometimes come to a halt, causing diseases such as Alzheimer’s.
The new findings, published in the November 2011 issue of the journal PLoS Biology, are also significant in that these molecular “locomotives” play a critical role in moving the chromosomes apart during cell division. Indeed, a number of the most important existing cancer drugs, such as taxanes and vinca alkaloids, work by targeting these transport systems in rapidly dividing cancer cells.
Kinesins are a family of microtubule motor proteins active in mitosis, or cell division. These kinesins use chemical energy from the hydrolysis of ATP (adenosine triphosphatase), a compound that the body produces from food to generate energy as needed by the body.
Using the Triton Resource, a medium-scale high performance computing system at UC San Diego’s San Diego Supercomputer Center (SDSC), researchers developed computational simulations of kinesin ‘engines’ hauling cargo along microtubule rails within cells. The findings show that electrostatic attraction between the engine and the rail is critically important in making the railway work. Videos showing the simulations can be viewed below:
“Precisely how kinesin motor proteins move along their microtubule tracks is an important question in biology,” according to J. Andrew McCammon, Joseph Mayer Chair of Theoretical Chemistry and Professor of Pharmacology at UC San Diego and a Howard Hughes Medical Institute Investigator, and senior author of the research paper, called Electrostatically Biased Binding of Kinesin to Microtubules. “We know that some kinesins have twin ‘heads’ that alternately bind to and step along microtubules in a coordinated walking action. But more usually, kinesins have only one head, and how single-headed kinesins produce force and movement is poorly understood.”
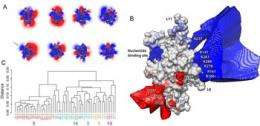
In the study, funded by the National Institutes of Health (NIH) and the National Science Foundation (NSF), researchers addressed this question and concluded that electrical attraction between single kinesin heads and microtubules is a critical factor deciding the direction of movement: each time the head approaches a microtubule, it slides forward by the electrical attraction between the engine and the track.
Implications for Nanoengineering
“This research shows that computational methods can be used to rationally design mutant molecular motors, with altered electrostatic properties, that can regulate the speed of the railway,” said researcher Barry J. Grant, now an assistant professor with the University of Michigan at Ann Arbor. Grant formerly was a member of McCammon’s research lab at UC San Diego.
In keeping with the train analogy, speeding up means having a more efficient transport of cargo, perhaps by means of a drug. Slowing the speed provides researchers with a good test of the general operational constraints for producing directed motion on the molecular scale, which is informative for future nanoengineering projects. Moreover, defects in motor-dependent processes, such as slowing down or stopping altogether, are associated with a large range of diseases, including neurodegeneration, tumorigenesis and developmental defects.
“Some of these calculations for these protein simulations required quite a large amount of computer memory, so the Triton Resource proved to be very helpful in this work,” said Grant, noting that each simulation in the project consumed about 7 gigabytes of ram/core with subsequent analysis of data sets measuring almost 2 terabytes in size. A single terabyte is one trillion bytes of data, about equal to the information printed on paper made from 50,000 trees.
“Ultimately, construction of molecular motors to arbitrary specifications will provide a powerful tool kit for therapeutic delivery and nanotechnology applications.” said McCammon.
Featuring a 2,000 processor computing cluster, a unique “large-memory” cluster for data-intensive computing, and a high capacity, high performance data storage system, the Triton Resource is offered on a space-available basis to researchers throughout the larger academic community, as well as private industry and government-funded organizations.
In addition to McCammon and Grant, other researchers contributing to the study include Dana M. Gheorghe, Robert A. Cross, and Maria Alonso from the Centre for Mechanochemical Cell Biology at the Warwick Medical School, University of Warwick in the U.K.; Wenjun Zheng, of the University of Buffalo, New York; Maciej Dlugosz, of the Interdisciplinary Centre for Mathematical and Computational Modelling at the University of Warsaw, Poland; and Gary Huber, with UC San Diego’s Department of Pharmacology.
Provided by University of California - San Diego