Researchers identify novel mechanism for regulation of gene expression
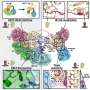
The Stowers Institute's Conaway Lab has demonstrated that an enzyme called Uch37 is kept in check when it is part of a human chromatin remodeling complex, INO80. The results were published in today's issue of Molecular Cell.
Uch37 is a "deubiquitinating enzyme" that can remove protein tags (called ubiquitin) from other proteins. The presence of one kind of ubiquitin tag on a protein can mark it for destruction, but others serve as marks to affect the activity of a protein. INO80 is a chromatin remodeling complex that is believed to function in both gene regulation and DNA repair by "unpacking" DNA from nucleosomes to allow access to chromosomal DNA.
Previously, the Conaway Lab demonstrated that Uch37 is associated with another multiprotein complex, the proteasome — a large protein complex that degrades unneeded or damaged proteins. In the new paper, the team shows that when bound to INO80, Uch37 can also be activated in the presence of proteasomes. Although the mechanism involved isn't totally clear, it seems to occur via a "touch and go" mechanism, in which proteasomes interact transiently with Uch37.
"Our findings suggest that activation of INO80-associated Uch37 by transient association of proteasomes with the INO80 complex could be one way proteasomes help to regulate gene expression," said Tingting Yao, Ph.D., Postdoctoral Research Fellow and lead author on the paper.
"Tingting's discovery of communication between INO80 and the proteasome provides new clues into the functions of both of these regulatory complexes," said Joan Conaway, Ph.D., Investigator and senior author on the paper. "In addition, it provides new insights into how deubiquitinating enzymes can be regulated — the ability to regulate these enzymes is very important because promiscuous removal of ubiquitin marks could lead to a failure to regulate properly the activities or levels of key enzymes and proteins in cells."
Source: Stowers Institute for Medical Research