New nanotechnology able to examine single molecules, aiding in determining gene expression
A new nanotechnology that can examine single molecules in order to determine gene expression, paving the way for scientists to more accurately examine single cancer cells, has been developed by an interdisciplinary team of researchers at UCLA's California Nanosystems Institute (CNSI), New York University's Courant Institute of Mathematical Sciences, and Veeco Instruments, a nanotechnology company.
Their work appears in the January issue of the journal Nanotechnology.
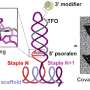
Previously, researchers have been able to determine gene expression using microarray technology or DNA sequencing. However, such processes could not effectively measure single gene transcripts—the building blocks of gene expression. With their new approach, the researchers of the work reported in Nanotechnology were able to isolate and identify individual transcript molecules—a sensitivity not achieved with earlier methods.
"Gene expression profiling is used widely in basic biological research and drug discovery," said Jason Reed of UCLA's Department of Chemistry and Biochemistry and the study's lead author. "Scientists have been hampered in their efforts to unlock the secrets of gene transcription in individual cells by the minute amount of material that must be analyzed. Nanotechnology allows us to push down to the level of individual transcript molecules."
"We are likely to see more of these kinds of highly multi-disciplinary research aimed at single molecule sequencing, genomics, epigenomic, and proteomic analysis in the future," added Bud Mishra, a professor of Computer Science, Mathematics, and Cell Biology from NYU's Courant Institute and School of Medicine. "The most exciting aspect of this approach is that as we understand how to intelligently combine various components of genomics, robotics, informatics, and nanotechnology—the so-called GRIN technology—the resulting systems will become simple, inexpensive, and commonplace."
Source: New York University