Novel virus breaks barriers between incompatible fungi
Scientists have identified a virus that can weaken the ability of a fungus to avoid pairing with other incompatible fungi, according to new research published in PLOS Pathogens. By promoting fungal pairing, the virus could aid transmission of additional unrelated viruses between fungi.
Fungi, like all other organisms, can recognize foreign substances; such non-self recognition can help protect against pathogens. Some fungi also use non-self recognition to avoid pairing and sharing genetic material with incompatible strains. The fungus Sclerotinia sclerotiorum, which infects hundreds of plant species worldwide, employs this strategy, which is known as vegetative incompatibility.
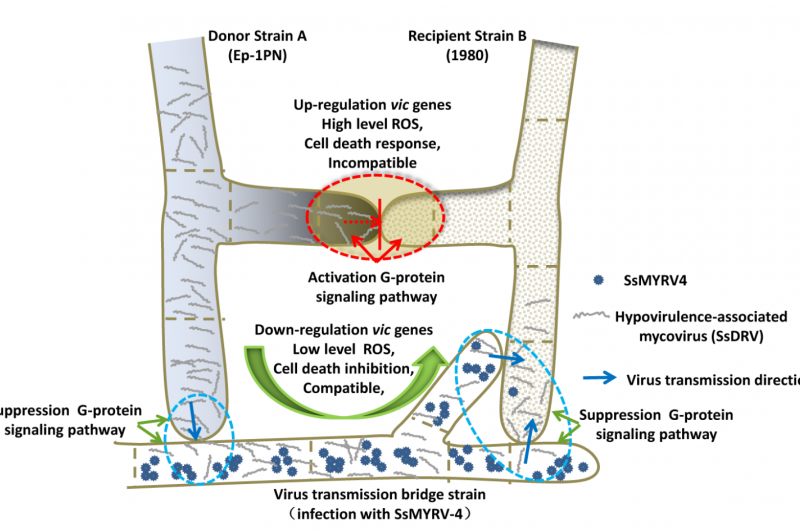
While studying S. sclerotiorum, Jiatao Xie of Huazhong Agricultural University, China, and colleagues discovered a virus they named Sclerotinia sclerotiorum mycoreovirus 4 (SsMYRV4). To better understand this novel virus, they grew infected S. sclerotiorum alongside other vegetatively incompatible strains and investigated the molecular effects.
The researchers found that SsMYRV4 decreased expression of S. sclerotiorum genes that promote vegetative incompatibility. Vegetative incompatibility is a molecular process that normally causes cell death when two incompatible strains touch each other; in this study, Xie's team found a reduction in the amount of cell death that normally occurs in intermingled colonies of incompatible strains.
S. sclerotiorum infected with SsMYRV4 successfully made connections with incompatible strains by fusing filamentous structures known as hyphae. To investigate the consequences, the scientists grew SsMYRV4-infected fungi alongside fungi infected with other unrelated viruses. They found that the unrelated viruses were able to pass through the fused hyphae, crossing between fungal pairs.
Vegetative incompatibility is considered a significant obstacle to using viruses to effectively control fungal diseases. These new findings could point to a new strategy that uses SsMYRV4 to weaken barriers between fungi. They could also improve understanding of virus ecology and evolution.
More information: Wu S, Cheng J, Fu Y, Chen T, Jiang D, Ghabrial SA, et al. (2017) Virus-mediated suppression of host non-self recognition facilitates horizontal transmission of heterologous viruses. PLoS Pathog 13(3): e1006234. DOI: 10.1371/journal.ppat.1006234
Journal information: PLoS Pathogens
Provided by Public Library of Science