January 31, 2012 feature
Something old, something new: Evolution and the structural divergence of duplicate genes
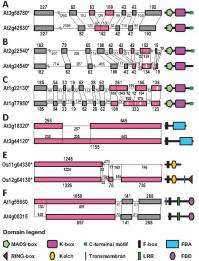
(PhysOrg.com) -- Gene duplications are arguably the driving force of organismal evolution – and if they survive, such duplicate genes will diverge in both regulatory and coding genomic regions. Coding divergences, in turn, can be caused by nucleotide substitutions or exon-intron structural changes. (Exons are DNA bases that are transcribed into mRNA and eventually code for amino acids in proteins. Introns are DNA bases found between exons, but which are not transcribed.) Scientists have had limited knowledge in the latter case until recently, when researchers at the Institute of Botany of the Chinese Academy of Sciences investigated structural divergences during the evolution of duplicate and nonduplicate genes. They found that such structural divergences are very common in duplicate gene evolution, and have resulted from three primary causes – exon/intron gain/loss, exonization/pseudoexonization (where an intronic or intergenic sequence becomes exonic, or vice versa), and insertion/deletion – each contributing differently to structural divergence. The scientists concluded that structural divergences play a more important role in the evolution of duplicate genes than nonduplicate genes.
The research, led by Professor Hongzhi Kong and Assistant Professor Guixia Xu in the Institute of Botany’s State Key Laboratory of Systematic and Evolutionary Botany, faced three main challenges in investigating the occurrence, importance and underlying mechanisms of structural divergences during the evolution of duplicate and nonduplicate genes.
“The first was to identify suitable duplicate genes for comparison,” Kong told PhysOrg.com. “Not all duplicate genes, albeit abundant, could be used for this purpose – if two genes have diverged too much, it would be difficult or even impossible to make a reliable comparison between them. The second,” Kong continues, “was to generate a reasonable alignment for each gene pair based upon which the underlying mechanisms for structural divergence were determined. The third was to calculate the genetic distance between genes, especially when changes in exon-intron structure have caused shifts in reading frame.”
Kong described the ways in which the team addressed these issues. “To identify suitable duplicate genes for this study, we only considered the most closely related duplicate genes – that is, sibling paralogs – simply because their evolutionary histories were relatively short and deducible. However, the problem with this strategy is that our estimates of structural divergence were somehow conservative. Nevertheless, because differences in exon-intron structure were widespread even between sibling paralogs, our results highlighted the prevalence and importance of structural divergence during duplicate gene evolution.“
To determine the underlying mechanisms for structural divergence, it is crucial to generate a reliable alignment for each paired sibling paralogs. “However,” Kong explains, “because such work relied heavily on the annotated gene structures, we first checked and evaluated the quality of gene annotation. We found that in plants, Arabidopsis and rice were the two species whose genomes have been most extensively and carefully annotated. We therefore focused exclusively on these species at this stage. We also found that in both Arabidopsis and rice, the annotations of some genes were likely better than others, simply because they play key roles in plant development and have been the focuses of functional studies. For this reason, and because of time and labor limits, we concentrated on seven well-known gene families.”
During alignment, the team also took into consideration alternative splicing to ensure that the observed differences in exon-intron structure were not the artifact caused by comparisons of transcription forms with different splicing choices. In other words, among the multiple transcription forms of the two paralogs, only those with the same splicing choices and the highest similarity were considered. “This is also a conservative strategy that further minimized the potential errors in gene annotation.” Kong adds.
Finally, says Kong, to calculate the genetic distances between genes, they only used the regions for which homology can be determined with confidence. “When structural changes have caused shifts in reading frame,” Kong points out, “corresponding regions were no longer homologous – especially when the corresponding amino acids were considered) and thus were excluded from further analyses. This was done manually for each of the investigated gene pairs.”
Kong also discussed the team’s conclusion that structural divergences have played a more important role during the evolution of duplicate than nonduplicate genes. “Many people believe that duplicate genes tend to evolve more rapidly than nonduplicate genes because of functional redundancy,” he observes. “However, in the past few decades, attention has been paid exclusively to nucleotide substitutions, possibly because they are easy to detect and investigate. Some people even believe that point mutation, especially those that can lead to replacements of amino acids with distinct biochemical properties, play overwhelming roles in gene evolution.”
Kong also points out that there are still scattered studies showing that changes in exon-intron structure have occurred and contributed to the generation of functionally distinct paralogs and orthologs (genes in different species that evolved from a common ancestral gene by speciation). “Actually, in many recent studies – especially those that focus on the evolution of multigene families – there are plenty of cases in which duplicate genes show obvious differences in exon-intron structure. This suggests that structural divergence have been widespread and important in gene evolution. Unfortunately, up until now, an extensive investigation of the prevalence, consequences and underlying mechanisms of structural divergence has been lacking.”
In other words, the group’s study is the first to deal with the general patterns of structural divergence in gene evolution. “The conclusion that structural divergence has played a more important role during the evolution of duplicate than nonduplicate genes will help understand why gene duplication has contributed greatly to the acquisition of novel physiological and morphological characters. Clearly, duplication and subsequent divergence of genes have led to the increase of the genetic and phenotypic diversity of life.”
In Kong’s opinion, their work will have at least three impacts. “Firstly, it highlights the importance of structural divergence in gene evolution, and may induce more broad and thorough studies on the other properties of structural divergence,” he explains. “This will help understand more about the general patterns of gene evolution.”
Secondly, he continues, it will help understand the possible defects or even errors of studies in which only EST, CDS or protein sequences were compared. “As I wrote in our paper,” he notes, “‘in the future, when two or more genes are compared, special attention should be paid to their genomic sequences. Without the knowledge of exon-intron organization, it is impossible to guarantee the reliability of the alignments of genes if structural divergences, especially those that can cause shifts of reading frame, have occurred.’"
Lastly, Kong says that their findings will stimulate reconsideration of some definitions now being widely used. “During the study, we feel that the differences or boundaries between many biological terms or concepts – such as alternative splicing and exonization/pseudoexonization, and exon/intron gain/loss and exon shuffling – are not very clear. We discussed this briefly in the paper, but more efforts are needed to clarify these issues.”
In terms of next steps in their research, Kong says that the team is pursuing in two directions. “One is to investigate the prevalence and underlying mechanisms of structural divergence in representative animals, such as humans and fruit flies, and yeasts to see whether structural divergence play equally important roles in these eukaryotic lineages. From our preliminary data, we’re rather certain that this is not always the case.”
Their other focus is to investigate many other properties of structural divergence. “For example,” Kong adds, “at present we have neither calculated the occurrence rates of each mechanism for structural divergence, nor have we known whether and to what extent natural selection has contributed to the process. Also, in my lab, we focus a bit more on the genetic and molecular basis for morphological evolution. We’ve found that duplication and diversification of a few regulatory genes – mostly transcription factor genes – are responsible for the alterations in floral characters. We’re also carrying out functional studies to see how changes in exon-intron structure have contributed to phenotypic evolution.”
Kong adds that their research is extremely laborious and time-consuming, because most steps have to be performed manually. “It would be great if automatic pipelines could be developed to speed up the process. There’s some software that accomplishes this, but for many reasons, the quality of the work is not always satisfactory. We’re currently collaborating with developers to improve the quality and speed of such applications.”
Beyond their own research in molecular evolution, genomics, and evolutionary developmental biology, Kong concludes, the team’s research findings may benefit any other areas that have connections with these fields.
More information: Divergence of duplicate genes in exon–intron structure. Published online before print January 9, 2012, PNAS January 24, 2012 vol. 109 no. 4 1187-1192, doi: 10.1073/pnas.1109047109
Copyright 2012 PhysOrg.com.
All rights reserved. This material may not be published, broadcast, rewritten or redistributed in whole or part without the express written permission of PhysOrg.com.