Shotgun sequencing finds nanoorganisms
For 11 years, Jill Banfield at the University of California, Berkeley, has collected and studied the microbes that slime the floors of mines and convert iron to acid, a common source of stream pollution around the world.
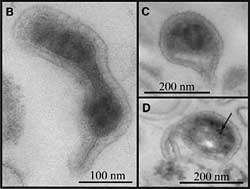
Imagine her surprise, then, when research scientist Brett Baker discovered three new microbes living amidst the bacteria she thought she knew well. All three were so small - the size of large viruses - as to be virtually invisible under a microscope, and belonged to a totally new phylum of Archaea, microorganisms that have been around for billions of years.
What made Baker's find possible was shotgun sequencing, a technique developed and made famous by Celera Corp., which used it to sequence the human genome in record time.
"It was amazing," said Banfield, a professor of earth and planetary science and of environmental science, policy and management at UC Berkeley since 2001. "These were totally new and very small organisms we didn't know how to culture with standard techniques. This shows the great promise of shotgun sequencing to profile a community of organisms without making any assumptions about what is there."
Nearly three years ago, Banfield employed shotgun sequencing to pick out the half-dozen bacteria and Archaea in the mine slime. It was the first successful example of community genomic analysis to profile the organisms in a drop of water - scummy water.
But Baker's discovery makes clear that shotgun sequencing can also pick out rare organisms too small to see easily, and too novel to be plucked out by other genetic techniques.
"We were essentially looking for new stuff, and we found it in all the samples studied, though at low abundance," said Baker, who is with the campus's Department of Earth and Planetary Science. "Shotgun sequencing is a better way to identify organisms than using other methods, like culturing or PCR (polymerase chain reaction), which can miss quite a lot of organisms."
Banfield, Baker and their UC Berkeley and University of Queensland, Australia, colleagues report their findings in the Dec. 22 issue of Science.
Banfield noted that the bacteria and newfound Archaea living in the highly acidic mine drainage are archetypes of the kind of life that could exist on other planets, such as in the iron- and sulfur-rich soil of Mars.
"This community of microbes is relevant to probing potential strategies for life on other planets, especially the life likely to exist on Mars," she said.
The organisms in the mine drainage, which live in a pink slick on pools of acidic green water, obtain energy by oxidizing iron - that is, generating rust -- and in the process create sulfuric acid and dissolve pyrite (iron sulfide or fool's gold) to release more iron and sulfur. This self-sustaining process creates the acidic drainage that pollutes creeks and rivers, including those around the researchers' study site, the Richmond Mine at Iron Mountain, Calif. The mine is one of the largest Superfund sites in the country.
Banfield has been trying to understand how the extremophiles - microbes that live in extreme environments - live together and generate the acid drainage that makes such mines toxic hazards. The green runoff from the mine, captured and treated by the Environmental Protection Agency, is a hot 108 degrees Fahrenheit, as acidic as battery acid, and loaded with toxic metals - zinc, iron, copper and arsenic.
In 2004, Banfield collaborated with the Department of Energy's Joint Genome Institute to shotgun sequence a drop of the slime. This type of sequencing involves homogenizing the organisms in the sample, isolating the combined DNA and breaking it into lots of random strands. Each strand is then sequenced, and a powerful computer is used to find overlaps so that the pieces can be properly reordered.
This process identified five separate genomes that corresponded to five bacteria and Archaea - four of them uncultivated at that time, though closely related to known microbes.
Baker probed the gene fragments more thoroughly to turn up three Archaea from a totally unknown group, probably representing a new phylum among the several dozen known phyla of Archaea. They fall within a large class of microbes known as thermophiles, which are Archaea that live in warm and even scalding conditions. Many of these thermophiles have been recovered from hydrothermal vents in the deep mid-ocean ridges, where lava boils up between continental plates.
Once Baker had found gene segments (ribosomal RNA) from three Archaea, he was able to fish the microbes out of the slime soup and found that they were extremely small, around 200 nanometers in diameter, the size of large viruses. Bacteria average about five times this diameter.
These therefore could be the smallest organisms ever found, though Baker needs to culture them before confirming this. Because they're so small, however, they may not be free-living.
"We're not sure they can live independently, whether they have enough genes to fend for themselves, but instead are symbiotic with another organism or are feeding off another organism," Baker said.
Baker now is trying to find the right conditions for these Archaea to thrive in a culture dish. For now, he has dubbed them ARMAN-1, -2 and -3, for Archaeal Richmond Mine Acidophilic Nanoorganisms. Ted Arman is the owner of the mine.
Source: University of California - Berkeley
















