Scientists Trace Corn Ancestry from Ancient Grass to Modern Crop
Researchers have identified corn genes that were preferentially selected by Native Americans during the course of the plant's domestication from its grassy relative, teosinte, (pronounced "tA-O-'sin-tE") to the single-stalked, large-eared plant we know today. The study revealed that of the 59,000 total genes in the corn genome, approximately 1,200 were preferentially targeted for selection during its domestication.
The study, by University of California, Irvine's Brandon Gaut and his colleagues, appears in the May 27 issue of the journal, Science.
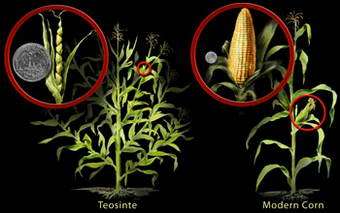
Image: Cultivated corn was domesticated from teosinte more than 6,000 years ago. During the process, corn lost the ability to survive in the wild, but gained valuable agricultural traits. The suppression of branching from the stalk resulted in a lower number of ears per plant but allows each ear to grow larger. The hard case around the kernel dissappeared over time. Today, we see just a few ears of corn growing on one unbranched stalk and enjoy larger ears covered with many, many rows of soft corn kernels. Credit: Nicolle Rager Fuller, National Science Foundation
Understandably, a primary goal of teosinte domestication was to improve the ear and its kernels. A teosinte ear is only 2 to 3 inches long with five to 12 kernels—compare that to corn's 12-inch ear that boasts 500 or more kernels! Teosinte kernels are also encased in a hard coating, allowing them to survive the digestive tracks of birds and grazing mammals for better dispersal in the wild. But, for humans, the tooth-cracking coating was undesirable so it was selectively reduced…and reduced…and reduced…until all that remains is the annoying bit of paper-thin, translucent tissue that sometimes sticks between the teeth when one munches corn on the cob.
To analyze the genes of modern corn and its ancestral teosinte, Gaut and his coworkers used relatively new genomic techniques to determine the DNA sequence of 700 gene bits in the two plants and used "population genetics," the study of genetic variation, to compare them.
"These results will provide important insights to modern corn breeders in their quest to establish hardier, higher-yielding corn plants," said Gaut. "The scientific approach will also be useful in the study of other domesticated organisms, plants and animals alike."
This work generally confirms the idea that corn went through a "population bottleneck," or a period when a significant portion of corn’s genetic diversity was lost, which typically marks a domestication event. Calculations using these data reveal that fewer than 3,500 teosinte plants may have contributed to the genetic diversity in modern corn.
Between 6,000 and 10,000 years ago, Native Americans living in what is now Mexico began domesticating teosinte, or the "grain of the gods," as the name has been interpreted to mean. Scientists cannot yet say how long this domestication process took, but they do know that around 4,500 years ago, a plant recognizable as today's corn was present across the Americas.
So, thousands of years before Gregor Mendel postulated his theories on genetics and heredity, indigenous Americans were breeding corn to select for desirable traits. By selectively breeding each generation, ancient farmers drastically changed teosinte's appearance, yield, grain quality and survivability—culminating in today's "corn." In fact, teosinte is so unlike modern corn, 19th century botanists did not even consider the two to be related.
"This is a very exciting finding," said Jane Silverthorne of the National Science Foundation's (NSF) biology directorate, which funded the project. "We are beginning to have a much clearer picture of what happened to the genes responsible for the structure of today’s corn plant."
A broad understanding of the genes present in modern-day corn will provide a foundation for improving it as well as its cousin cereal crops. Target goals include yield increases, improved insect and pathogen resistance, enhanced environmental adaptability, and improved nutritional value. To that end, sequencing the entire genome of corn is also critical to improving the crop and its value in human subsistence. To that end, sequencing the entire genome of corn is also critical to improving the crop and its value in human subsistence.
According to the U.S. Department of Agriculture (USDA), nearly 12 billion bushels of corn were harvested in the United States in 2004, which will be used for a diverse array of products including livestock feed, ethanol and plastic consumer items, as well as food. The National Corn Growers Association reported that 2003 corn exports were valued at $4.5 billion.
Supported by NSF's Plant Genome Research Program, this collaborative project included Gaut and co-workers at the University of California, Irvine, together with scientists from the USDA-Agricultural Research Service, the University of Missouri and the University of Wisconsin. NSF is part of an interagency program along with the U.S. Department of Energy and the USDA that plans to support the sequencing of the corn genome over the next three years.
Source: NSF