March 27, 2015 feature
Seeing the (UV) light: Previously undetected difference in human mutation rate unique to Europeans
Stuart Mason Dambrot
guest writer
(Phys.org)—Although humans are a single species, not all genetic variation is shared between populations – and the ability to sequence our entire genome has allowed scientists to catalogue mutations that occur in one ethnic group alone. These so-called population-private mutations give researchers a unique window into recent human history. Recently, graduate student Kelley Harris – a scientist at University of California, Berkeley – revealed a previously undetected difference between Europeans and other ethnic groups by comparing population-private mutation frequencies from Europe, Asia, and Africa, finding that Europeans experience higher rates of a specific mutation type that has known associations with UV light exposure. Harris concludes that while it is unclear whether the excess mutations are harmful or directly related to the UV sensitivity of light skin, her results demonstrate that the human mutation rate has evolved on a much faster timescale than previously believed, with implications for cancer genetics, anthropology and other fields of inquiry.
Harris discussed the paper she published in Proceedings of the National Academy of Sciences with Phys.org, first addressing the challenge of revealing a previously undetected difference between Europeans and other ethnic groups contrasting frequencies of mutations private to Europe, Asia, and Africa. "A question that's pretty hot in human genetics right now is whether the human mutation rate has changed over time. The idea that motivated me to do this project is that there are many different ways that DNA can mutate (an A base can change to G, a C to A, and so on), and I reasoned that if some biochemical force had changed the mutation rate at some point during human history, it was likely to have affected certain mutation types more than others," Harris tells Phys.org. However, she adds, while she assumed that if a difference did exist it would be subtle, she was surprised to find such a large difference between the mutational spectra of different populations. In fact, she notes that the pattern described in her paper was observed in the results of the first simple test she performed (although additional effort was needed to show that this pattern was valid and not simply a genome sequencing artifact). "There are hundreds of cryptic data quality problems that can affect genome sequences, and it was challenging to show that so many potential sources of error weren't creating the illusion of a biologically significant finding."
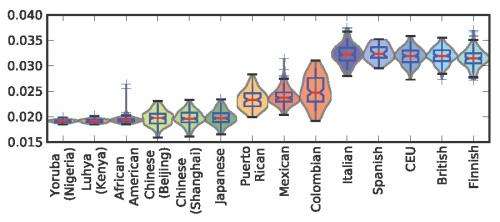
A more specific challenge was determining that Europeans experience higher rates of a specific mutation type that has known associations with UV light exposure. "When I performed a broad screening test for mutation frequency differences between populations," Harris explains, "the excess of the mutation type TCC->TTC in Europe immediately jumped out as hugely significant." The TCC->TTC mutation is the most common somatic mutation present in melanoma skin cancers, as well as the mutation most frequently induced in vitro by UV. "I didn't know at the time that this mutation type had associations with UV exposure, but these associations turned up pretty quickly in the course of further online research."
The paper also described how Harris had to account for the possibility that mutation rates might have changed owing to recent regional adaptations affecting DNA repair. "The possibility of local adaptation affecting DNA repair wasn't so much a challenge as a motivating factor for doing the analysis," Harris says, adding that her strategy was to describe how real genetic variation differs from idealized models of mutation as a simple process that doesn't change over time. "That being said, while I was really excited to find evidence that DNA replication fidelity appears to differ between populations, we're still a long way from knowing the mechanism behind this difference or whether it affects human health."
One question that Harris hopes will generate some excitement among experimentalists who know how to grow yeast or culture cells concerns the prospects of determining that folate depletion could cause some of the mutations observed in UV-irradiated cells, and that these same mutations might appear in the germ line of a light-skinned individual rendered folate-deficient by sun exposure. "I'd love to be able to compare nutrient-deficient cell populations to control cell populations and test whether cells deprived of folate or other DNA synthesis cofactors experience different patterns of mutation than cells grown without nutrient deficiencies." She points out that while counting new mutations requires a lot of DNA sequencing, but this sort of experiment should become cheaper and more feasible as the cost of DNA sequencing continues to decline.
The paper also notes that differences in metabolic rate can drive latitudinal gradients in the rate of molecular evolution. "Several previous studies observed that organisms with high metabolic rates appear on average to have higher mutation rates than organisms with low metabolic rate. I'm not sure if those observations have any relationship to my observations about humans, since they deal with much more highly diverged organisms that have much more metabolic rate diversity than exists within the human species, but it would be really interesting if that were the case."
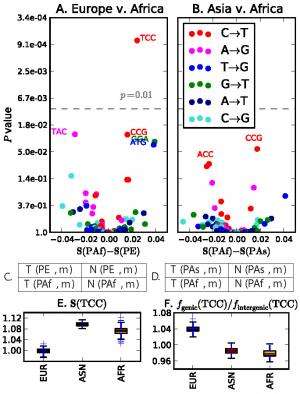
![Overrepresentation of 5’-TCC-3’ → 5’-TTC-3’ within Europe. (A and B) The x coordinate of each point in gives the fold frequency difference (fPE(m) - fPAf(m)/fPAf(m)) [respectively (fPAS(m) - fPAf(m))/ fPAf(m))], and the y coordinate gives the Pearson’s χ2 P value of its significance. Outlier points are labeled with the ancestral state of the mutant nucleotide flanked by two neighboring bases, and the color of the point specifies the ancestral and derived alleles of the mutant site. (C and D) The χ2 contingency tables used to compute the respective P values in A and B. (E) The distribution of f(TCC) across bins of 1,000 consecutive population-private SNPs. Only chromosome-wide frequencies are shown for chromosome Y because of its low SNP count. Credit: Harris K (2015) Evidence for recent, population-specific evolution of the human mutation rate. Proc Natl Acad Sci USA 112:11 3439-3444. Overrepresentation of 5’-TCC-3’ → 5’-TTC-3’ within Europe](https://scx1.b-cdn.net/csz/news/800a/2015/overrepresen.jpg)
Harris explains an idea from the scientific literature that the hypothesized hominoid evolutionary slowdown1 might be related to lengthening generation time. "It's important to know the rate per year at which mutations accumulate to use DNA to answer questions related to human history," Harris tells Phys.org. To estimate this rate, she explains, researchers usually compute the fraction of sites at which two DNA sequences differ and divide this by an estimate of how long ago the sequences shared a common ancestor. However, she points out that although this estimation procedure seems straightforward, mutation rate estimates tend to be lower when they're based on differences between parents and children than those between a human sequence and a chimp sequence. "One possible explanation for this is that the mutation rate was higher at the time closer to that when the human/chimp common ancestor lived."(This phenomenon is referred to as the hominoid slowdown because it postulates that the mutation rate slowed down as hominoids evolved into anatomically modern humans.)
"The reason this might be a consequence of lengthening generation time," she says, "is that mutations occur most often during cell divisions – and the older humans are when they reproduce, the more cell divisions occur along the lineage between when one human child is conceived and when his or her own child is eventually conceived ."
That said, Harris continues, the hominoid slowdown is just one hypothesis to explain the observed discrepancy between different human mutation rate estimates; another posits that DNA sequencing errors interfere with accurately estimating the small numbers of differences between parents and offspring. "Since DNA sequencing quality issues potentially make it so difficult to count mutations and estimate their absolute rates, I decided to try an indirect approach to shed light on whether the human mutation rate has recently changed, reasoning that if some populations had higher frequencies of specific mutation types than other populations, it probably indicated that the absolute rates of those mutation types had recently changed – a more parsimonious explanation than a zero-sum event where some mutation types increased in rate and other mutation types simultaneously decreased in rate. Using this approach, I was able to infer that the mutation rate has been evolving without relying on any possibly inaccurate numerical rate estimates."
Relatedly, Harris' approach presents a caveat to the standard molecular clock2 assumption – the idea that mutations accumulate over time at a constant rate, like clock ticks. "This supposition predicts that the number of differences between two DNA sequences is linearly proportional to the amount of time they have been diverging since their last common ancestor. The molecular clock assumption is usually assumed to be false when you're comparing really distantly related organisms like humans and flies, since DNA replication and error correction has had a lot of evolutionary time to change, but it's usually assumed to be true when comparing organisms as close together as humans and chimps. When you assume that the molecular clock assumption is true and that you know the mutation rate per year, you have a lot of power to use DNA sequences to infer historical dates. This is one reason that my finding of mutation rate change in Europeans is significant: To some extent, it contradicts an assumption that human geneticists usually take for granted – and that's important for applying genetics to anthropology."
There are also the apparent differences in the distribution of de novo mutations between human and chimpanzee male and female germ lines, as well as between different mutation types. Regarding the human/chimpanzee differences, Harris notes that this is an observation from the scientific literature. "A recent study at Oxford3 sequenced families of chimps and found that many more mutations were transmitted from fathers to children than from mothers to children. This makes sense given that sperm undergo many more cell divisions than do eggs, and thus accumulate mutations faster. This same trend is thought to hold in humans – but in humans, the difference between male and female mutation rates appears less extreme."
Harris also refers to the Oxford study when discussing differences in the distribution of de novo mutations between different mutation types, pointing out that a high percentage of the mutations they found in the chimp family they sequenced were so-called CpG mutations – that is, a mutation in which a C nucleotide separated by a phosphate (p) from, and located next to, a G nucleotide, to a T nucleotide. "This mutation type has a different chemical mechanism than do most mutations where a C changes to a T," Harris explains. "In most organisms, C nucleotides located next to G nucleotides have higher mutation rates than most sites in the genome, and this difference in mutability looks like it might be more extreme within chimp genomes than within human genomes. This is an example of a scenario where one mutation type – in this case, CG->TG – might have increased in rate along the chimp lineage relative to the rates of other mutation types."
Harris has several plans for her continued and future research, one being to analyze whether the mutagenic spectrum might have changed numerous times during great ape evolution and beforehand given that it was able to change during the last 60,000 years of human history. "This is a question I hope to address by applying the techniques I developed in this paper to new datasets. For example," she illustrates, "a large dataset of great ape genomes was published last year, and I'm looking forward to searching for differences between the mutation spectra of great ape species the same way I looked for differences between human populations for this paper. This should give us a better idea of whether the observed difference between Europeans and Africans/Asians was a real anomaly that had functional significance, or whether it was just one of many random mutation rate changes that are more frequent than we've assumed them to be." Harris also plans to investigate DNA from anatomically modern humans who died thousands of years ago, since an increasing number of these ancient DNA sequences are starting to be produced at high quality and released for public use.
Another goal in Harris' calendar is pinpointing finer-scale mutation rate changes and other avenues of research. "The statistical analysis techniques I used in this paper are really simple – I used the same chi-square test taught in high school biology class because the signal of TCC->TTC mutations being more frequent in Europeans than in other groups is so strong that more sophisticated techniques weren't needed to detect it. However, now that I know that the mutational process can differ between human populations, I'm going to develop more robust statistical tools that will be intended to have higher power to detect interpopulation differentiation signals that might be weaker than the dominant signal I studied in this paper, but still be real and thereby important." A key question she'd like to answer is how frequently and routinely mutational processes do change – and she sees detecting smaller differences between populations as important step toward achieving that.
In addition, Harris in interested in determining whether the TCC->TTC excess in Europeans is a trait with a simple genetic basis – that is, if it might be the result of a causal mutation in a single gene associated with DNA replication or repair. "Scientists have mapped the locations of a lot of genes that are associated with variation in traits like height or disease risk, and knowing the genetic basis of the variation in some trait can help you test whether or not that variation was likely driven by natural selection," Harris tells Phys.org. "Usually, we think about a person's DNA sequence encoding a trait that's external to the DNA – but my question is unusual in that I'm interested in a trait that is itself a feature of the DNA sequence." Harris acknowledges that this intertwining presents some technical challenges, but states that if she can develop new techniques for overcoming those challenges, those techniques could allow researchers to study variations in other traits that are endogenous, rather than exogenous, to DNA.
In closing, Harris notes that other areas of research might benefit from her study. "Cancer genetics is an area that I've recently started reading about because of the association of melanoma with the mutation type TCC->TTC that jumped out in the results of my study. Although I'm far from being an expert on cancer genetics at this point, I'm hopeful that that field could have something to gain from a better description of how mutational processes can vary in a benign way between human populations."
Written for you by our author Stuart Mason Dambrot—this article is the result of careful human work. We rely on readers like you to keep independent science journalism alive. If this reporting matters to you, please consider a donation (especially monthly). You'll get an ad-free account as a thank-you.
Publication details
Evidence for recent, population-specific evolution of the human mutation rate, Proceedings of the National Academy of Sciences (2015) 112:11 3439-3444, doi:10.1073/pnas.1418652112
Related:
1Genomic data support the hominoid slowdown and an Early Oligocene estimate for the hominoid–cercopithecoid divergence, Proceedings of the National Academy of Sciences (2004) 101(49):17021–17026, 10.1073/pnas.0407270101
2The molecular clock runs more slowly in man than in apes and monkeys, Nature (1987) 326:93-96, doi:10.1038/326093a0
3Strong male bias drives germline mutation in chimpanzees. Science (2014) 344(6189):1272–1275, doi:10.1126/science.344.6189.1272
Journal information: Proceedings of the National Academy of Sciences , Nature , Science
© 2015 Phys.org














