Researchers 'unzip' molecules to measure interactions keeping DNA packed in cells
(PhysOrg.com) -- Anyone who has ever battled a stuck zipper knows it's a good idea to see what's stuck, where and how badly -- and then to pull hard. A Cornell research team's experiments involve the "unzipping" of single DNA molecules. By mapping the hiccups, stoppages and forces along the way, they have gained new insight into how genes are packed and expressed within cells.
The research, "High-resolution dynamic mapping of histone-DNA interactions in a nucleosome," published online Jan. 11, 2009, in Nature Structural and Molecular Biology, was led by Michelle Wang, associate professor of physics and Howard Hughes Medical Institute Investigator. Collaborators on the project included physics graduate student Michael Hall and John Lis, the Barbara McClintock Professor of Molecular Biology and Genetics.
DNA - the molecules that contain genetic information - are nucleic acids often illustrated as long, thin strands of double helices. DNA fits inside cell nuclei by being wound like thread around proteins called histones, forming tightly packed bundles called nucleosomes. But that same DNA must often be uncoiled and accessed by such enzymes as RNA polymerase, which the researchers liken to a motor because it moves along the DNA in the process of gene transcription.
"There is this paradox," Lis explained. "On one hand you need compaction and the packing away of DNA. On the other hand, you need accessibility, so the cellular machines can read the information contained in the DNA."
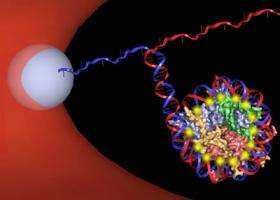
Trying to understand what happens during that unwrapping process is at the heart of this research team's efforts. By unzipping each DNA double helix through a nucleosome using an optical trap -- a technique developed in Wang's lab -- they unwrapped strands of DNA from their histone cores, observing, with near-base pair accuracy, the interactions that took place along the way.
"Our hope is that if we can establish and understand the interactions within the nucleosome, we can begin to understand how the motor proteins can invade the nucleosome," Wang said.
Optical trapping involves a focused beam of light that can "trap" small objects. A refractive sphere is chemically attached to the DNA strand, and the optical trap moves the sphere, allowing the researchers to unzip the DNA strands apart by pulling, Hall explained. By doing so, the researchers re-created what happens in the cell when DNA uncoils from the histone core, and they measured the blips along the way -- for example, when the DNA strand had to be pulled apart from a protein molecule -- and how much force was needed to keep going.
"It's really like a zipper," Hall said. "And when there is a protein in there, it's kind of like you have a piece of cloth stuck. You know you can get it out, but you just have to pull harder, and then it pops out. That's basically the same way we can detect where the interactions are with the proteins."
The researchers have performed the first direct, precise measurements of histone-DNA interactions. Their findings could help uncover how changes to the histones or DNA sequences affect how motor proteins access genetic information in cells.
"If we have that knowledge, we can extrapolate that information to apply to different scenarios and different motor motions," Wang said.
Reference: Nature Structural and Molecular Biology, doi: 10.1038/nsmb.1526
Provided by Cornell University