Researchers use new approach to predict protein function
In a paper published online this month in the journal Nature Chemical Biology, researchers report that they have developed a way to determine the function of some of the hundreds of thousands of proteins for which amino acid sequence data are available, but whose structure and function remain unknown.
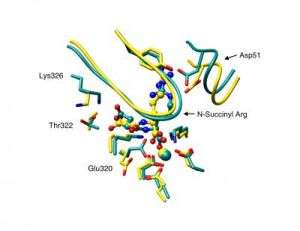
The research team, led by University of Illinois biochemistry professor John A. Gerlt, is the first to use a computational approach to accurately predict a protein’s function from its amino acid sequence. Their “in silico” (computer-aided) predictions were validated in the laboratory by means of enzyme assays and X-ray crystallography.
The new approach involved searching databases of known proteins for those with amino acid sequences that had the greatest homology to the unknown proteins. The researchers then used the three-dimensional structures of the most closely matched known proteins in their analyses of protein function.
Using the structural data obtained from this homology modeling, the team performed computerized docking experiments to quickly evaluate whether the unknown proteins were likely to bind to any of a vast library of potential target molecules, or substrates. Determining which substrate binds to a given protein is vital to understanding the protein’s function.
“This study describes an integrated approach using experimental techniques, computational techniques and X-ray crystallography for predicting the function of a protein of previously unknown function,” Gerlt said.
These methods will speed the task of identifying the biological roles of some of the hundreds of thousands of proteins whose functions have not yet been discovered.
“Rather than trying to do (laboratory) experiments on 30,000 compounds to determine if they are substrates, with this approach you might do experiments on 10,” Gerlt said.
The study involved a family of proteins within the large and diverse enolase superfamily. Enolases are enzymes that catalyze the breakdown of glucose and related compounds into other molecules as needed for metabolism.
The enzymes within the enolase superfamily utilize similar reaction mechanisms to one another but catalyze different reactions, complicating the task of discovering their function. There are more than 3,000 proteins in the enolase superfamily, and a majority of them have not yet been fully – or accurately – characterized. (The new study also revealed that one family of enolase proteins had been misclassified.)
Gerlt and his colleagues expect that the computational approach they pioneered will help scientists more efficiently tackle the problem of understanding these – and other – unknown proteins.
“There are 4 1/2 million protein sequences in the sequence databanks, and maybe the functions are known for, can be assigned to, half of those with some reliability,” Gerlt said. “That tells you that there is a lot of biology to be discovered.”
Source: University of Illinois at Urbana-Champaign















