A molecular map for aging in mice
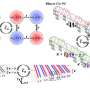
Researchers at the National Institute of Aging and Stanford University have used gene arrays to identify genes whose activity changes with age in 16 different mouse tissues. The study, published November 30 in PLoS Genetics, uses a newly available database called AGEMAP to document the process of aging in mice at the molecular level. The work describes how aging affects different tissues in mice, and ultimately could help explain why lifespan is limited to just two years in mice.
As an organism ages, most tissues change their structure (for example, muscle tissues become weaker and have slow twitch rather than fast twitch fibers), and all tissues are subject to cellular damage that accumulates with age. Both changes in tissues and cellular damage lead to changes in gene expression, and thus probing which genes change expression in old age can lead to insights about the process of aging itself.
Previous studies have studied gene expression changes during aging in just one tissue. The new work stands out because it is much larger and more complete, including aging data for 16 different tissues and containing over 5.5 million expression measurements.
One noteworthy result is that some tissues (such as the thymus, eyes and lung) show large changes in which genes are active in old age whereas other tissues (such as liver and cerebrum) show little or none, suggesting that different tissues may degenerate to different degrees in old mice.
Another insight is that there are three distinct patterns of aging, and that tissues can be grouped according to which aging pathway they take. This result indicates that there are three different clocks for aging that may or may not change synchronously, and that an old animal may be a mixture of tissues affected by each of the different aging clocks.
Finally, the report compares aging in mice to aging in humans. Several aging pathways were found to be the same, and these could be interesting because they are relevant to human aging and can also be scientifically studied in mice.
CITATION: Zahn JM, Poosala S, Owen AB, Ingram DK, Lustig A, et al. (2007) AGEMAP: A gene expression database for aging in mice. PLoS Genet 3(11): e201. doi:10.1371/journal.pgen.0030201, www.plosgenetics.org
Source: Public Library of Science