DNA of prehistoric cave bears sequenced
The genomic DNA sequencing of an extinct Pleistocene cave bear species – the kind of stuff once reserved for science fiction – has been logged into scientific literature thanks to investigators from the U.S. Department of Energy (DOE) Joint Genome Institute (JGI). The study, published in the June 2 online edition of the journal Science, has set the research community’s sights on traveling back in time through the vehicle of DNA sequencing to reveal the story of other extinct species – including our nearest relatives, the Neandertals.
Until now, researchers have been stymied in attempting to sequence genomes of extinct species. The DOE JGI scientists overcame many of the difficulties normally associated with recovery of DNA from ancient samples. DNA starts degrading at death when microbes attack the decaying carcass to utilize the nutrients present in the dead organism as an energy source. What remains – and confounds efforts to sequence and characterize these artifacts – is an overabundance of microbial contaminants, along with the occasional DNA fingerprints contributed unwittingly by modern fossil hunters or lab workers.
“Among the limitations of previous ancient DNA studies was that they were restricted to mitochondrial DNA sequences,” said Eddy Rubin, DOE JGI director, in whose laboratory the work was conducted. “While mitochondria are great for learning about evolutionary relationships between species, to understand the functional differences between extinct and modern species we really need genomic DNA, and nobody has been able to purify and sequence large quantities of DNA from these old samples.
“We applied the standard techniques that we normally use for modern genome projects to ancient DNA specimens – a brute-force, high-throughput sequencing approach,” Rubin said. “With this strategy, we weren’t terribly concerned that much of what we were sequencing were microbial contaminants, but hoped that among the large amounts of sequence that we were generating would be some of the ancient DNA we really were interested in.
“We were looking for the proverbial needle in the haystack,” Rubin said. “It worked – among the expected lion’s share of contaminants we recovered reasonable amounts of 40,000-year-old cave bear DNA and useful information from it. We were lucky in that we had a very powerful magnet, in the form of industrial-strength computing, to tease out the interesting data from a hodgepodge of different DNAs.”
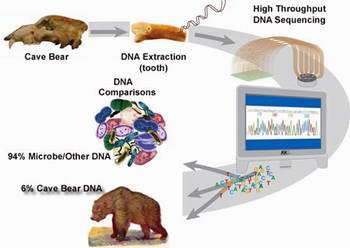
It turned out that about 6 percent of the sequence from the sample yielded cave bear sequence; the rest represented a mosaic of microbial contaminants. Nevertheless, within that fraction was a range of genomic sequence types, including fragments of 21 genes identified by comparing the cave bear samples to the complete dog genome sequence that exists in public databases. Dogs and bears, which diverged some 50 million years ago, are 92 percent similar on the sequence level.
The samples of cave bear bones and teeth from the study were collected from two cave locations in Austria. Extinct for more than 10,000 years, these particular cave bears, Ursus spelaeus, whose remains are found in abundance, were related to the ancestors of modern brown bears and polar bears. Fossil and cave-painting evidence shows that ancient humans interacted with the cave bears.
“When people hear about our success, they immediately think about how this strategy could work for dinosaurs,” Rubin said. But barring some fundamental misunderstanding about the nature of DNA decay, he says it is highly unlikely that viable DNA will be recovered from 150- to 200-million-year-old Jurassic age samples.
Currently, the theoretical age limit is about 100,000 years for samples preserved in the same conditions that the cave bear specimens were found – relatively dry, high altitude, with moderate temperatures – perhaps longer if frozen.
“We picked cave bear as an initial test-case ancient DNA target because the samples we used in the study are roughly the same age as Neandertals,” Rubin said. “Our real interest is in hominids, which include humans and the extinct Neandertal – the only other hominid species that we have to compare with humans.
“Our nearest living relative is the chimp, and that’s five million years of divergence,” he said. “Although we are very similar on a sequence level, there are obvious phenotypic differences. Next, we would like to access and evaluate genomic information about other hominid species, Neandertals in particular, as they represent probably our closest prehistoric relative.”
Another possibility is to apply the techniques to the remains found recently in Indonesia of the Flores Man, nicknamed “the Hobbit.” Although younger, at 18,000 years old, the skeletons were found in a tropical environment that may have accelerated the DNA degradation process. The Flores Man is believed to have derived from a more ancient hominid lineage, Homo erectus, which diverged from modern humans some two million years ago – as compared to Neandertal, which diverged 500,000 years ago.
The approach would be to optimize the protocols that succeeded in the cave bear project to extract useful quantities of DNA from Neandertal and Flores Man, so that two hominid species could be compared to humans. Then, by including chimps in that comparison, Rubin said, researchers would gain insight into the genomic changes between chimps and hominids and help describe the chain of evolutionary events that led to humans.
“This research represents the merging of two previously unrelated fields – ancient species investigations and powerful high-throughput DNA sequencing technology, DOE JGI’s forte,” said Dr. Aristides Patrinos, DOE’s associate director of science for Biological and Environmental Research, whose office supported the published work. “This is yet another of the many advances enabled by the original investment made by DOE in the Human Genome Project.”
First author on the Science paper was Lawrence Berkeley National Laboratory postdoctoral fellow James Noonan, who joined DOE JGI’s Chris Detter and Doug Smith, and collaborators at the Max Planck Institute for Evolutionary Anthropology, Leipzig, Germany and the Institute of Paleontology, University of Vienna, Vienna, Austria.
Source:DOE Joint Genome Institute