Researcher Fashions DNA to Further Advances in Nanotechnology

In the fifty-year history since the structure of DNA was first revealed, what was once a Nobel prize- winning research discovery has become an omnipresent cultural icon co-opted for promoting everything from fragrances to musical acts. Now, the familiar DNA double helix is serving as a microscopic trellis in order to further advances in nanotechnology aimed at improving human health.
Image: An atomic force microscopy image of the thrombin/DNA complex. The DNA appears as two long threads in the center of the image, with the brighter spots corresponding to thrombin proteins attached to the DNA.
Hao Yan, a researcher at the Biodesign Institute at Arizona State University and an assistant professor in ASU's Department of Chemistry and Biochemistry, recently created unique arrays of proteins tethered onto self-assembled DNA nanostructures.
While other efforts in recent years have focused on learning how to build DNA-based nanostructures, Yan's work is novel because it makes it feasible to attach any desired biomolecule onto DNA nanostructures. Such work is an important step and can serve as a future foundation for biocatalytic networks, drug discovery or ultrasensitive detection systems.
"Rationally-designed DNA nanoscale architectural motifs have for a long time been envisioned as scaffolds for directing the assembly of biomolecules such as proteins into a functional network," said Yan. "However, the methods to control such assemblies are still scarce. A robust and modular approach is needed. "
In his results, Yan and fellow institute researchers Yan Liu, Chenxiang Lin, and Hanying Li have taken advantage of the base pairing properties of DNA to make the DNA nanostructures. By controlling the exact position and location of the chemical bases within a synthetic replica of DNA, Yan could potentially fashion a variety of DNA assemblies.
In this case, Yan created a triple crossover DNA tile, consisting of three side-by-side helices just six nanometers in width and 17 nanometers in length. One nanometer is one-billionth of a meter. By programming into the assembly a short sequence of DNA that recognizes a particular protein, called an aptamer, Yan created a DNA molecule that could now function as a biomolecular tether.
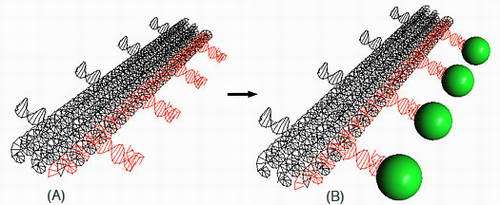
Image: A 3-D model of a DNA nanostructure. In figure A, three long cylinders of individual helices (black) contain regularly spaced intervals of aptamers (red) which can bind to a protein. In figure (B), a thrombin protein (green circle) binds to the DNA aptamer structure.
"This is the first time ever an aptamer has been utilized to link proteins to self-assembled DNA nanoarrays," said Yan.
Yan integrated an aptamer that recognizes the protein thrombin, which is an important protein vital to blood clotting. The technique allows for Yan to precisely control both the position and spacing of the thrombin proteins on the DNA nanoarray.
Yan's confirmed his results by using atomic force microscopy, where the thrombin proteins bound to the DNA nanoarray are seen as beads on a string. Because of the ability of the protein binding to be visualized, one intriguing application of the technique may be in the application toward single molecule proteomics studies.
"We are actively discussing applying this technology to single molecule proteomics and to study protein-protein interactions because the distance between interacting proteins could be controlled with nanometer accuracy," said Yan.
Also, by attaching different proteins onto the DNA scaffold, Yan could directly visualize the binding of a drug to its target molecule or recreate metabolic pathways on a single array to mimic the way different organelles function in a cell.
The article was recently published early online for the journal Angewandte Chemie and can be found at dx.doi.org/10.1002/anie.200501089
Source: Arizona State University















