Technique provides snapshot of all genes in human genome
(PhysOrg.com) -- Like Silly Putty lifting an imprint of the Sunday comics off a newspaper, complex enzymes called RNA polymerases lift information off DNA strands. The polymerases then transcribe those genetic instructions onto RNA for making proteins that cells use for basic functions.
In the Dec. 19 issue of Science, Cornell researchers report on a new technique that takes a snapshot of all the locations on the human genome where RNA polymerases actively transcribe genes. The method provides a new and highly sensitive way to pinpoint all the active and silent genes in the human genome.
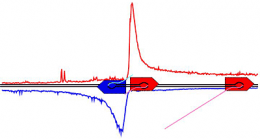
The researchers also report on a new mechanism: Scientists always thought that RNA polymerases read DNA in one direction, by starting at a bit of DNA at the front of a gene -- the so-called promoter -- and moving to the end of the gene. But the new polymerase maps reveal that polymerases also appear on the other side of the promoter and run in the opposite direction.
"We always thought that polymerases followed one direction, but now we have polymerases going both ways," said John Lis, the paper's senior author and the Barbara McClintock Professor of Molecular Biology and Genetics at Cornell.
The polymerases going forward follow through to the end of the gene and transcribe the information for making proteins, but for reasons yet unknown, polymerases going in the opposite direction travel only a short distance and then stop. Researchers hypothesize that these opposite-facing polymerases could be holding open a segment of DNA for transcription.
The researchers also observed that the transcription process pauses because polymerases accumulate near the promoter before moving to the end of the gene. The pause may have a structural function, the researchers suggest, to hold open the site for other polymerases to enter. The pause seems to occur at genes that respond to signals (called regulated genes) and could allow time to prepare the gene for a rapid and coordinated response to a signal. Or, a pause may be necessary to make sure that all the necessary components are in place for proper transcription.
"A pause can likely be serving a different role at different genes," said Josh Waterfall, a postdoctoral researcher in Lis' lab and a co-author of the paper. "It's only in the last few years that experiments suggest these pauses are a common pattern on a lot of genes." The new study confirms that the high frequency of locations where polymerases build up, as seen in other studies, are sites where transcription pauses.
Leighton Core, a graduate student in Lis' lab and the paper's lead author, developed the new mapping technique by modifying an older technique. "The assay is actually around 30 years old, but it traditionally only measured the polymerases locations at small discrete units," he said. "But we were able to adapt the assay to measure actively engaged polymerases across the human genome."
This paper is one of four simultaneous papers appearing in Science that discuss these divergent polymerases for the first time. The papers have been online at Science Express since Dec. 4.
Provided by Cornell University