Researchers Tackling Unsolved Questions About Protein Structures
A University of Arizona research team is exploring the evolutionary origins of protein structures. Their findings will help people better understand how proteins evolved to carry out the instructions encoded in the genes of every living thing.
Protein molecules are made up of chains of amino acids. These chains bend and fold into a dizzying array of three-dimensional shapes and structures, depending on the order of the amino acids in a given chain. Those varied structures are part of what allow the proteins – which are assembled based on instructions coded in DNA – to regulate everything from an organism's growth and metabolism to the ways messages are transmitted from cell to cell. Protein structures are at the heart of how organisms function.
However, the evolution of those structures is still poorly understood, because there are few observed examples of proteins that have clearly evolved from one shape to another.
"The origin of the diversity of protein structures is a major unsolved problem," explains Matthew H.J. Cordes, a member of the UA's BIO5 Institute and an associate professor of biochemistry and molecular biophysics.
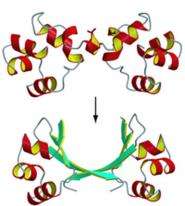
Cordes' lab is solving that problem. Two graduate students in his lab, Christian M. Roessler and Branwen M. Hall, have located protein molecules in two different viruses that have dramatically different structures: One protein has a helical, or corkscrew, shape, while the other is shaped more like a hairpin. Yet these very different proteins have similar amino acid sequences and perform similar functions – binding to DNA to help the viruses replicate and spread – making it likely that they had a common ancestor.
"Somehow, mutations converted the corkscrew structure to the hairpin structure," Cordes says of the finding, which was recently reported in the Proceedings of the National Academy of Sciences.
While this isn't the first example of structural differences among proteins with a common ancestor, it may be the most dramatic natural example of related proteins retaining clear similarity in amino acid sequence while undergoing major reorganization of their structure.
"This finding strongly confirms that evolutionary processes produce new protein shapes," Cordes says. "It could become a textbook example of the reality and beauty of evolutionary changes in structure." He adds that some proteins in this family with the hairpin shape bind to DNA more strongly than those with the corkscrew shape, though it is too early to tell if this is always the case. It's not yet known whether such an advantage helped drive the hairpin structure's evolution.
Cordes' graduate students found their protein pair via an unusual method: Roessler and Hall used a stepping-stone technique to make a series of small "jumps" among closely related proteins, following minute structural changes from one protein to another until they "landed" at a protein that was dramatically different from the one they'd started with, yet was still related to it.
Cordes' lab is now working out the details of the specific mutations that might have caused their two proteins to diverge from one another. They also plan to use their stepping-stone technique to shed light on the evolutionary links among other proteins.
"This is like space exploration," Cordes says. "We're journeying through the protein universe, step by step."
Source: University of Arizona

















