Illuminating Study Reveals How Plants Respond to Light
Most of us take it for granted that plants respond to light by growing, flowering and straining towards the light, and we never wonder just how plants manage to do so. But the ordinary, everyday responses of plants to light are deceptively complex, and much about them has long stumped scientists.
Now, a new study "has significantly advanced our understanding of how plant responses to light are regulated, and perhaps even how such responses evolved," says Michael Mishkind, a program director at the National Science Foundation (NSF). This study, which was funded by NSF, will be published in the November 23, 2007 issue of Science.
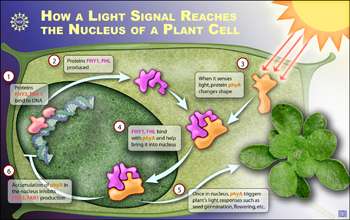
By conducting experiments with Arabidopsis--a small flowering plant widely used as a model organism--the researchers discovered that the plant prepares to respond to light while it is still in the dark, even before it is exposed to light. This preparation involves producing a pair of closely related proteins (known as FHY3 and FAR1) that increase production of another pair of closely related proteins (known as FHY1 and FHL) that had been identified in previous studies as critical participants in the plant's light response.
Haiyang Wang, a member of the research team from Boyce Thompson Institute for Plant Research, says that the plant probably stockpiles these proteins needed for light responses in the dark for the same reason that a traveler fills his car's gas tank the night before a morning journey: in order to be able to get going, without delay, at first light.
With a plant so primed in the dark, it detects and responds to light via the following steps:
-- Light-sensing pigment proteins known as phytochrome A located in the cytoplasm of plants cells detect the light in the far-red end of the spectrum.
-- The phytochrome A is activated through a change in shape that allows it to bind to FHY1 and FHL.
-- The binding of FHY1 and FHL to phytochrome A results in the accumulation of phytochrome A in the cell nucleus, possibly by helping to import phytochrome A into the nucleus.
-- The activated phytochrome A changes the activity of genes located in the cell nucleus that govern plant growth and development.
-- Resulting changes in gene expression produce the plant's developmental responses to light, such as growth, flowering and straining towards the light.
Although these steps had been identified in previous studies, the discovery of how FHY3 and FAR1 regulate plant responses to light adds an important new dimension to our understanding of them.
Moreover, the researchers also discovered the existence of a negative feedback loop between accumulations of phytochrome A in the cell nucleus and the FHY3 and FAR1 proteins that prime the plant's light response system: the more phytochrome A accumulates in the nucleus, the less FHY3 and FAR1 proteins are produced, and so less phytochrome A is imported into the nucleus. "This feedback loop serves as a built-in brake that limits the flow of light responses," says Wang.
"I can't explain why nature created such a complex process to trigger a plant's light responses," says Wang with a sigh. Among the process's complexities is a resemblance between FHY3 and FAR1 proteins and certain enzymes produced by some mobile DNA elements or so-called "jumping genes." (Jumping genes are so named because they can move between various positions in a cell's genetic code.) "This resemblance initially puzzled the research team when we were trying to identify the molecular function of the proteins," says Wang.
Nevertheless, the resemblance between the FHY3 and FAR1 proteins and jumping gene enzymes may represent a biological blessing in disguise. Why? Because the researchers now believe that they have built a convincing case that FHY3 and FAR1 may have evolved from the jumping gene material. If indeed the proteins did so, this important chapter in evolution may have helped make possible the establishment of flowering plants on earth, says Wang.
Source: NSF

















