New model could improve some drugs' effectiveness
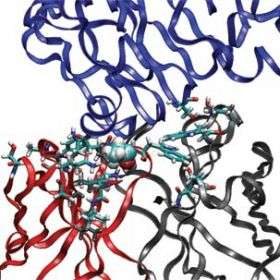
MIT researchers have developed a computer modeling approach that could improve a class of drugs based on antibodies, molecules key to the immune system. The model can predict structural changes in an antibody that will improve its effectiveness.
The team has already used the model to create a new version of cetuximab, a drug commonly used to treat colorectal cancer, that binds to its target with 10 times greater affinity than the original molecule.
The work, which will appear Sept. 23 in an advance publication of Nature Biotechnology, results from a collaboration using both laboratory experiments and computer simulations, between MIT Professors Dane Wittrup and Bruce Tidor.
“New and better methods for improving antibody development represent critical technologies for medicine and biotechnology,” says Wittrup, who holds appointments in MIT's Department of Biological Engineering and Department of Chemical Engineering. Tidor holds appointments in Biological Engineering and the Department of Electrical Engineering and Computer Science.
Antibodies, which are part of nature's own defense system against pathogens, are often used for diagnostics and therapeutics. Starting with a specific antibody, the MIT model looks at many possible amino-acid substitutions that could occur in the antibody. It then calculates which substitutions would result in a structure that would form a stronger interaction with the target.
“Combining information about protein (antibody) structure with calculations that address the underlying atomic interactions allows us to make rational choices about which changes should be made to a protein to improve its function,” said Shaun Lippow, lead author of the Nature Biotechnology paper.
“Protein modeling can reduce the cost of developing antibody-based drugs,” Lippow added, “as well as enable the design of additional protein-based products such as enzymes for the conversion of biomass to fuel.” Lippow conducted the research as part of his thesis work in chemical engineering at MIT, and is now a member of the protein engineering group at Codon Devices in Cambridge, Mass.
“Making drugs out of huge, complicated molecules like antibodies is incredibly hard,” said Janna Wehrle, who oversees computational biology grants at the National Institute of General Medical Sciences, which partially supported the research. “Dr. Tidor's new computational method can predict which changes in an antibody will make it work better, allowing chemists to focus their efforts on the most promising candidates. This is a perfect example of how modern computing can be harnessed to speed up the development of new drugs.”
Traditionally, researchers have developed antibody-based drugs using an evolutionary approach. They remove antibodies from mice and further evolve them in the laboratory, screening for improved efficacy. This can lead to improved binding affinities but the process is time-consuming, and it restricts the control that researchers have over the design of antibodies.
In contrast, the MIT computational approach can quickly calculate a huge number of possible antibody variants and conformations, and predict the molecules' binding affinity for their targets based on the interactions that occur between atoms.
Using the new approach, researchers can predict the effectiveness of mutations that might never arise by natural evolution.
“The work demonstrates that by building on the physics underlying biological molecules, you can engineer improvements in a very precise way,” said Tidor.
Expanding on that theme, Wittrup and Tidor also co-teach a class and are writing a textbook focusing on connecting fundamental molecular and cellular events to biological function through the use of mathematical models and computer simulations.
The team also used the model with an anti-lysozyme antibody called D44.1, and they were able to achieve a 140-fold improvement in its binding affinity. The authors expect the model will be useful with other antibodies as well.
Source: Massachusetts Institute of Technology















