Researchers Report Ability to Detect Cancer at Earliest, Curable Stage
Researchers at the Moores Cancer Center at the University of California, San Diego report that they have developed a new method for detecting cancer very early in its development, when it consists of just a few cells. The best existing detection methods are not able to detect a tumor until it consists of about one million cells.
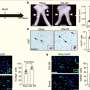
The paper*, published in the April 18 issue of the online journal PLoS ONE, describes a series of proof-of-concept experiments in which the researchers, working with two cancer cell lines, were able to select out and amplify tiny amounts of cancer-causing DNA in the presence of more than 99.9 percent of normal DNA. Current methods for identifying deleted DNA would not work in clinical settings because they require isolation of relatively pure cancer cells. This is not feasible for clinical samples, which typically contain large amounts of the person’s normal cells.
“We have developed a new technology for very early detection of virtually any type of solid-tumor cancer based upon damaged DNA, which is where all cancers begin,” said co-author Dennis A. Carson, M.D., professor of medicine and director of the Moores Cancer Center. “We are now working with engineers toward the fabrication of the clinical devices that will enable this to be widely used in patients.”
Carson said they are several years away from clinical testing, but ultimately individuals will be able to be screened for DNA markers of cancer cells using simple clinical samples such as blood or urine. Using this same technology, physicians will be able to easily and inexpensively monitor the status of patients by looking for the DNA markers. If the treatment worked, there would be no mutated DNA and the patient would be cured. Such monitoring would also shorten the time needed to determine if the treatment is not working so another approach could be instituted.
The technology, called Primer Approximation Multiplex PCR (PAMP), is based upon an enzyme reaction that only works when a piece of DNA has been deleted or is abnormally joined to another piece of DNA, according to co-author Yu-Tsueng Liu, M.D., Ph.D., assistant project scientist and director of the biomarker laboratory at the Moores Cancer Center. The exact location of the mutation does not matter. The method will detect any mutated DNA in the presence of normal DNA, and amplify only the mutant molecules.
Liu explains: “When a cancer cell mutates, it often brings together two pieces of DNA that are normally apart. We have developed an enzyme reaction that works well only when two DNA pieces that are normally separated are close together. This technology amplifies the mutant DNA and then uses a microarray to identify the specific mutation. Our experiments were conducted on a specific gene mutation that is well-known for its role in cancer, called CDKN2A, but this technology would work on any DNA abnormality.”
Source: UCSD