Scientists have discovered that mutations in a gene known as SOS1 account for many cases of Noonan syndrome (NS), a common childhood genetic disorder which occurs in one in 1,000-2,500 live births. NS is characterized by short stature, facial abnormalities, and learning disabilities, as well as heart problems and predisposition to leukemia.
Led by researchers at Harvard Medical School-Partners Healthcare Center for Genetics and Genomics (HPCGG) and Beth Israel Deaconess Medical Center (BIDMC), the findings are reported in the December issue of Nature Genetics, which appears on-line today.
"Noonan syndrome is the most common single gene cause of congenital heart disease," explains co-senior author Benjamin Neel, MD, PhD, Director of the Division of Cancer Biology at BIDMC and professor of medicine at Harvard Medical School (HMS).
"Although previous work had identified mutations in the PTPN11 gene as the cause of Noonan syndrome in nearly 50 percent of cases [and mutations in an oncogene known as KRAS in a small subset of severe cases] the identity of the gene or genes responsible for fully half the cases had not been elucidated," Neel said.
To identify candidate genes, a group led by HMS instructor Amy Roberts, MD, and director of HPCGG Raju Kucherlapati, PhD, conducted genetic analysis of over 100 children with Noonan syndrome. This large cohort of NS patients had neither PTPN11 nor KRAS mutations.
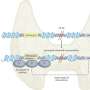
"From this group, we identified SOS1 mutations in approximately 20 percent of the cases," explains Kucherlapati, the Paul C. Cabot professor of genetics at HMS. After modeling the positions of the mutations on crystal structures of SOS1, the scientists made recombinant versions of the mutants and expressed them in mammalian cells, where it was discovered that they promoted excessive activation of RAS and its downstream target, MAP kinase, the same pathway activated by PTPN11 mutations.
"These results are the first example of activating mutations in an exchange factor in human disease," notes Neel, explaining that for families at risk for Noonan syndrome, the findings will aid in prenatal diagnosis and genetic counseling for the disorder.
"Furthermore," Neel adds, "because the other two genes that cause Noonan syndrome are also mutated in several types of leukemia and solid tumors, our findings may also expand our knowledge of cancer pathways."
Source: Beth Israel Deaconess Medical Center