Protein folding: Building a strong foundation
Like a 1950's Detroit automaker, it appears that nature prefers to build its proteins around a solid, sturdy chassis.
A new study combining advanced computational modeling and cutting-edge experiments by molecular biologists at Rice University and Baylor College of Medicine suggests that the most stable parts of a protein are also the parts that fold first.
The findings appear in the Sept. 13 issue of the journal Structure.
Nature refuses to choose between form and function when it comes to protein folding; each protein's function is directly related to its shape, and when proteins misfold – something that's known to occur in a number of diseases like Alzheimer's and Huntington's – they don't function as they should.
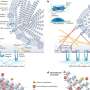
In the new study, scientists designed and tested a new computational approach that aimed to study proteins with known shapes in order to ascertain which of their parts were the most stable in the face of chemical and thermal fluctuations.
"We found that the most stable parts of the final, folded protein come together first during the folding," said co-author Pernilla Wittung-Stafshede, associate professor of biochemistry and cell biology and of chemistry. She said the findings could help both scientists who are attempting to design synthetic proteins with a particular shape and scientists who are attempting to associate the shape and function of naturally occurring proteins.
The computational approach tested in the experiment was developed by the research group of co-author Jianpeng Ma, associate professor of bioengineering at Rice and associate professor of biochemistry and molecular biology at Baylor College of Medicine. Ma's group, which mainly focuses on multi-scale protein structure modeling and prediction, developed highly accurate knowledge-based potential functions that made the current collaborative study possible.
"As far as we know, no one has ever used this type of knowledge-based, statistical approach to predict the stability cores of proteins," Ma said. "Our results suggest that thermodynamics and kinetics are closely correlated in proteins and appear to have co-evolved for optimizing both the folding rate and the stability of proteins."
Wittung-Stafshede's group, which specializes in experimental studies related to protein folding, tested the model's predictions against experimental data gathered for several forms of the protein azurin, a copper-containing protein that folds into a sandwich-like structure called a beta sheet, which is a common fold in nature that consists of two beta-sheets of amino-acid strands meshed together.
"In folding study the right combination of expertise in computational and experimental approaches is vital for success," Ma said. "Our collaborative team has set an excellent example for future study."
Source: Rice University