Theory shows mechanism behind delayed development of antibiotic resistance
Inhibiting the "drug efflux pumps" in bacteria, which function as their defence mechanisms against antibiotics, can mask the effect of mutations that have led to resistance in the form of low-affinity drug binding to target molecules in the cell. This is shown by researchers at Uppsala University in a new study that can provide clues to how the development of resistance to antibiotics in bacteria can be delayed.
The introduction of antibiotics as drugs in the treatment of bacterial infections in the post-WWII years was a revolutionized medicine, and dramatically improved the health condition on a global scale. Now, 60 years later, growing antibiotic resistance among pathogens has heavily depleted the arsenal of entailed effective antibiotic drugs.
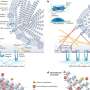
Antibiotics function by attacking vital molecules in bacteria. Bacteria, in turn, protect themselves either by using "drug efflux pumps" for antibiotics or through mutations that reduce the binding of the antibiotic to its target molecules inside the bacteria cell. Through these changes, bacteria develop resistance to antibiotics.
The new study is published in the journal Proceedings of the National Academy of Sciences in the US. Professor Mľns Ehrenberg's research team at Uppsala University has shown experimentally and theoretically explained how the inhibition of these drug efflux pumps can completely mask the resistance effect of mutations that reduce the affinity of antibiotics to their target molecules in the bacteria cell. The effect of the mutations is entirely hidden when the pumps are unable to remove the antibiotic sufficiently quickly in relation to the dilution of the antibiotic through cell growth and cell division.
"This masking effect can provide clues to how the development of resistance to antibiotics in bacteria can be delayed," says Mľns Ehrenberg.
The study introduces a new tool for understanding and delaying the development of resistance in bacteria.
Source: Uppsala University (news : web)