Mouse model highlights histone methylation as distinguishing feature for leukemia subtypes
Research using a new mouse model has led to the identification of a potential therapeutic target for a type of leukemia commonly associated with an unfavorable prognosis. The study, published by Cell Press in the November issue of the journal Cancer Cell, also validates examination of histone modification as a strategy for distinguishing cancer subtypes.
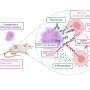
MLL rearrangements have been observed in some cases of acute lymphoblastic leukemia (ALL) and acute myeloid leukemia (AML). ALL patients with rearranged MLL have a relatively poor prognosis when compared with patients who have other types of ALL. Although several mouse models of MML-rearranged AML, modeling MLL-rearranges ALL has proven difficult. Recent studies have suggested that some MLL fusion proteins enhance gene expression by recruiting the histone H3 lysine79 (H3K79) methytransferase, DOT1L. Addition of a methyl group to histones plays an important regulatory role in the control of gene expression and is an example of an epigenetic change, a chemical alteration that does not change the DNA sequence.
"Aberrant recruitment of DOT1L to the promoters of MLL target genes may be a common feature of many oncogenic MLL fusion proteins. However, the extent of H3K79 methylation changes and the specificity of these epigenetic changes for MLL-rearranged leukemias have not been defined," says senior study author Dr. Scott A. Armstrong from the Dana-Farber Cancer Institute, Children's Hospital and Harvard Medical School.
Dr. Armstrong and colleagues created a mouse model in which conditional expression of MLL-AF4 induced both ALL and AML. They demonstrated that their model faithfully recapitulated human ALL resulting from MLL-AF4 translocation and identified a critical role for H3K79 methylation in MLL-AF4-driven gene expression and transformation. Importantly, suppression of DOT1L resulted in a reduction of methylated H3K79 and expression of critical MLL-AF4 target genes. "Our findings support the inhibition of DOT1L as a potential therapeutic approach in this disease and that this mouse model should be useful for assessment of therapeutic approaches for MLL-rearranged ALL," offers Dr. Armstrong.
"Our study also demonstrates that widespread differences in an epigenetic histone modification can distinguish different cancer subtypes and malignant versus normal cells," explains Dr. Armstrong. "Inasmuch as epigenetic modifications are likely to be linked to the maintenance of gene expression in many human malignancies, these strategies may be applicable to a wide variety of cancer subtypes."
Source: Cell Press