Abnormal 'editing' of gene messages may be a cause of lupus
Researchers at Wake Forest University have uncovered evidence that the abnormal “editing” of gene messages in a type of white blood cell may be behind the development of lupus. Scientists hope the finding will lead to earlier diagnosis, a way to monitor patients’ response to therapy and possibly a new way to treat the disease.
The findings, reported online in the journal Immunology, involve an enzyme that “edits” and modifies the messages of genes before the protein-making process. It is protein molecules that carry out the instructions of our genes and determine how an organism looks, how well its body metabolizes food or fights infection, and even how it behaves.
Dama Laxminarayana, Ph.D., assistant professor of internal medicine and senior author, said that in systemic lupus erythematosus, the normal editing process goes awry, causing a shift in the balance of proteins that results in impaired functions in T cells, a type of white blood cell involved in the regulation of immune functions.
Impaired T cell function is a hallmark of lupus, a complex chronic autoimmune disorder that can range from a benign skin disorder to severe, life-threatening multisystem disease. It primarily affects women in the child-bearing years and is more common in blacks.
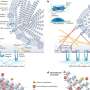
The current research, which involved 13 patients with lupus and eight healthy participants, was based on Laxminarayana’s earlier findings that 150-kDa ADAR1, one of the three enzymes involved in editing gene messages, is higher in the T cells of lupus patients compared to those without lupus. ADARs are ademosine deaminases that act on RNA.
Laxminarayana made the initial finding about 150-kDa ADAR1 levels in 2002 and has been working to solve the mystery of how it is related to the development of lupus. In the current study, Laxminarayana found that the higher levels of 150-kDa ADAR1 alters the editing induced by two other ADAR enzymes and may cause an imbalance of proteins. Editing by the two other ADAR enzymes is a normal cellular process; it is 150-kDa ADAR1 that causes normal editing to go awry.
The process is complicated and took Laxminarayana years to uncover. The current studies demonstrate that, essentially, too much 150-kDa ADAR1 results in an increase in the gene message of Phosphodiesterase 8A1 (PDE8A1), which is involved in the disruption of normal cell signaling and impairing cell function.
“150-kDa ADAR1 is the culprit,” Laxminarayana said. “We are now working to find a safe way to block it.”
In addition to targeting the enzyme as a treatment strategy, Laxminarayana said 150-kDa ADAR1 could also be used as a biomarker to detect the disease earlier, to monitor how patients respond to therapy, and to measure disease intensity.
Source: Wake Forest University