Genetics of imatinib resistance in acute lymphoblastic leukemia
In the September 15th issue of Genes & Development, Drs. Richard T. Williams, Willem den Besten, and Charles J. Sherr at Howard Hughes Medical Institute, St. Jude Children’s Research Hospital in Memphis TN, lend new insights into how an aggressive form of acute lymphoblastic leukemia (ALL) develops, and how sensitivity to the targeted chemotherapeutic drug, imatinib, can be diminished through interactions between tumor cells and the host microenvironment.
ALL, a cancer of the bone marrow affecting 4,000 US residents annually, is characterized by the over-production of immature white blood cells. An aggressive form of ALL results from a chromosomal translocation, known as the Philadelphia chromosome (Ph), in which segments from chromosomes 9 and 22 are aberrantly fused together. Ph+ ALL is far more prevalent in adults (~30% of adult ALL) than in children (~4% of pediatric ALL), but it carries a poor prognosis in both age groups. Ph+ cells express a protein (encoded by an oncogene created by the chromosome fusion) called BCR-ABL. BCR-ABL is a constitutively active enzyme, a tyrosine kinase, which promotes uncontrolled cell proliferation.
Continuous treatment with the BCR-ABL tyrosine kinase inhibitor, imatinib, has revolutionized the therapy of another form of Ph+ cancer, chronic myelogenous leukemia (CML), by inducing durable remissions. However, the response of Ph+ ALL patients is not nearly as good, leading to shorter remissions and more rapid emergence of imatinib resistance. In general, Ph+ CML and ALL patients that fail imatinib therapy develop mutations in the BCR-ABL kinase that make them drug-resistant, but the reasons underlying the increased rate of emergence of mutant clones in Ph+ ALL has not been satisfactorily explained.
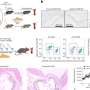
Williams and colleagues tracked the development of imatinib resistance, using a mouse model of Ph+ ALL. They engineered BCR-ABL-expressing lymphocyte progenitors that also lack the tumor suppressor protein ARF (which is deleted in more than 30% of Ph+ ALL patients, but not in CML patients, at their time of diagnosis). Interestingly, ARF-deficient lymphocytes expressing BCR-ABL were so highly aggressive that inoculation of as few as 20 such cells into healthy mice induced fatal ALL in less than 3 weeks. “Although experiments with CML support the concept that these leukemias arise from a rare population of ‘cancer stem cells’, our work on Ph+ ALL emphasizes that this need not be the case,” says Williams.
Further genetic experiments revealed that signals from the bone marrow micro-environment of the host animals were able to sustain the viability of ARF-deficient leukemia cells in the face of imatinib therapy. “We suspect that similar signals may nurture ARF-deficient Ph+ ALL cells in patients,” says Sherr, “thereby allowing the rapid emergence of imatinib-resistant clones.”
Source: Cold Spring Harbor Lab