Taking 'Chips' to the Next Level of Gene Hunting
Researchers at the Johns Hopkins’ High Throughput Biology Center have invented two new gene “chip” technologies that can be used to help identify otherwise elusive disease-causing mutations in the 97 percent of the genome long believed to be “junk.”
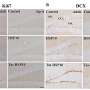
A variety of DNA microarray technology, one of the two new chips, called the TIP-chip (transposable element insertion point) can locate in the genome where so-called jumping genes have landed and disrupted normal gene function. This chip is described online this week in the Proceedings of the National Academy of Sciences.
The most commonly used gene chips are glass slides that have arrayed on them neat grids of tiny dots containing small sequences of only hand-selected non-junk DNA. TIP-chips contain on them all DNA sequences. Because each chip can hold thousands of these dots - even a whole genome’s worth of information - scientists in the future may be able to rapidly and efficiently identify, by comparing a DNA sample from a patient with the DNA on the chip, exactly where mutations lie.
“With standard chips, we’re missing a big piece of the picture of mutations in humans because they look only at the meaty parts of genes, but the human genome is only 3 percent meaty parts,” says Jef Boeke, Ph.D., Sc.D, professor of molecular biology and genetics and director of the HiT (High Throughput Biology Center), who spearheaded both studies at the Institute of Basic Biomedical Sciences at Hopkins. “The other 97 percent also can contain disease-causing mutations and is often systematically ignored,” he says.
Boeke and his team have focused particularly on transposable elements, segments of DNA that hop around from chromosome to chromosome. These elements can, depending on where they land, wrongly turn on or off nearby genes, interrupt a gene by lodging in the middle of it, or cause chromosomes to break. Transposable elements long have been suspected of playing a role vital to disease-causing mutations in people. Boeke hopes that the TIP-chip eventually can be used to look for such mutations in people.
The new TIP-chip contains evenly sized fragments of the yeast genome arrayed in dots left to right in the same order as they appear on the chromosome. Boeke’s team used the one-celled yeast genome as starting material because, unlike the human genome, which contains hundreds of thousands of transposable elements of which perhaps a few hundred are actively moving around, the yeast genome contains only a few dozen copies.
Like a word-find puzzle, where words are hidden in a jumbled grid of letters, the TIP-chip highlights exactly where along the DNA sequence these elements have landed. By chopping up the DNA, amplifying the DNA next to the transposable elements and then applying these amplified copies to the TIP chip, the researchers were able to map more than 94 percent of the transposable elements to their exact chromosome locations.
The second new chip, described in a separate report published in the Nov. 3 issue of Nature Methods, contains twice the amount of genetic information of current DNA chips.
“This one lets us look at twice as much as we could in the past, which means essentially that all chip experiments become faster and cheaper and can be done on an ever larger scale,” says Boeke. The chips his team currently uses cost about $400 per experiment. If the amount of information can be quadrupled, “it would be four experiments for the price of one,” he says.
Standard chips contain one layer of DNA dots that read from left to right, like the across section of a crossword puzzle. Boeke’s new double-capacity chips hold two layers of dots, a bottom layer that reads across and a top layer that reads down, again using the crossword analogy. So if their experiment lights up a horizontal row of dots, the researchers learn that the data maps to the region of the genome contained in the bottom layer; likewise, if the experiment highlights a vertical row, the data correspond to the top layer.
Says Boeke, “It’s so easy to differentiate the top and bottom layers. Next we’re going to try adding another layer reading diagonally” to triple the amount of genomic information packed onto the tiny chips.
Source: Johns Hopkins Medical Institutions