Study finds potential cell receptors to reduce antibiotic resistance
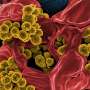
The bacterium Pseudomonas aeruginosa is the leading cause of hospital-acquired infections. The pathogen is resistant to many antibiotics so treating those infections, particularly in patients with compromised immune systems, is difficult.
A new study from UT has identified certain chemical receptors in cells that could deceive the bacteria and improve patient response to drugs.
The study was published this week in the Proceedings of the National Academy of Sciences.
Igor Zhulin, the study's lead author, is a joint faculty professor at the UT-Oak Ridge National Laboratory Joint Institute for Computational Sciences. He is based in the UT Department of Microbiology. Co-authors include Davi Ortega and Aaron Fleetwood, former UT graduate students who now are at the California Institute of Technology and the UT Health Sciences Center in Memphis, respectively.
A grant from the National Institutes of Health supported the study.
Antibiotic resistance occurs when bacteria change in ways that reduce or eliminate the effectiveness of drugs, helping the bacteria survive, continue to multiply, and cause more harm.
Researchers have been seeking to develop drugs that will, in effect, trick the bacteria rather than kill them, because bacteria often become resistant to antibiotics that kill, Zhulin said.
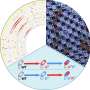
He and his research group study bacterial receptors and signaling pathways—the communication process that governs cell activities and coordinates cell actions. The pathways receive information from receptors and adjust cell functions accordingly.
In the Pseudomonas aeruginosa cell, there are 26 chemoreceptors—sensory units that respond to chemical stimuli by collecting information about the environment and feeding it to four signaling pathways, which then control cellular responses.
Zhulin and his group discovered that by comparing the protein sequences of different bacterial receptors and looking for specific patterns using computers, they could identify how 23 of those receptors shared the same unique amino acid patterns. The three other receptors had individually different patterns.
They concluded that to deceive the Pseudomonas aeruginosa cell, one strategy would be to feed misinformation to its chemoreceptors.
The work is one step toward helping scientists adapt antibiotics to better attack infections.
"This study will help in choosing novel targets and designing strategies for potential new antibiotics design," Zhulin said.
He added that if a future drug aims at preventing the pathogen from moving across surfaces of a cell as part of the infection process, then a single protein—the chemoreceptor feeding information into the pathway controlling surface movement—can be targeted.
More information: Davi R. Ortega et al. Assigning chemoreceptors to chemosensory pathways inPseudomonas aeruginosa, Proceedings of the National Academy of Sciences (2017). DOI: 10.1073/pnas.1708842114
Journal information: Proceedings of the National Academy of Sciences
Provided by University of Tennessee at Knoxville