This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Researchers present an unsupervised learning-based optical fiber imaging system
Fiber-optic imaging methods enable in vivo imaging deep inside hollow organs or tissues that are otherwise inaccessible to free-space optical techniques, playing a vital role in clinical practice and fundamental research, such as endoscopic diagnosis and deep-brain imaging.
Recently, supervised learning-based fiber-optic imaging methods have gained popularity due to their superior performance in recovering high-fidelity images from fiber-delivered degraded images or even scrambled speckle patterns. Despite their success, these methods are fundamentally limited by their requirements for strictly-paired labeling and large training datasets.
The demanding training data requirements result in time-consuming data acquisition, complicated experimental design, and tedious system calibration processes, making it challenging to satisfy practical application needs.
In a recent publication in Light: Science & Applications, Dr. Jian Zhao from the Picower Institute for Learning and Memory at the Massachusetts Institute of Technology, Dr. Xiaowen Hu and Dr. Axel Schülzgen from the College of Optics and Photonics (CREOL) at the University of Central Florida, and their colleagues presented an unsupervised learning-based optical fiber imaging system.
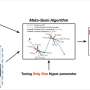
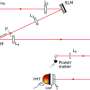
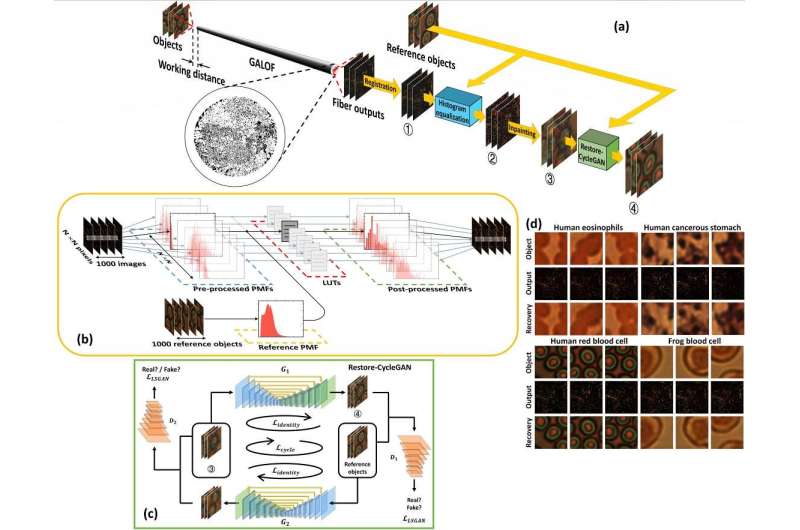
This system integrates a customized Cycle Generative Adversarial Network (CyleGAN), named Restore-CycleGAN, with Glass-Air Anderson Localizing Optical Fiber (GALOF). The application of Restore-CycleGAN removes the restrictions of labeled training data, yet maintains high-quality imaging recovery, while the unique physical properties of the GALOF's modes support high-fidelity and highly robust imaging processes and guarantee the successful implementation of unpaired imaging training.
Due to the mutual promotion between the learning algorithm and optical devices, the Restore-CycleGAN-GALOF method achieves nearly artifact-free and robust transport of full-color biological images through a meter-long optical fiber using a simple one-shot training process with a small training dataset of only 1000 image pairs, without requiring paired training imaging data. The training data size is reduced by about ten times compared to previously reported supervised learning methods.
The Restore-CycleGAN-GALOF method demonstrated high-fidelity, full-color image transport capabilities for various biological samples, including human and frog blood cells, human eosinophils, and human stomach cancer cells, under both transmission and reflection imaging modes.
Furthermore, this imaging process exhibited resilience against 60-degree strong mechanical fiber bending and large working distance variations of up to 6 millimeters. Remarkably, the Restore-CycleGAN-GALOF method produced high-accuracy predictions for test data that were never included in the training process, indicating strong generalization in the small data regime.
Despite the Restore-CycleGAN-GALOF's superior performance, the system design and experimental process are relatively simple. The scientists summarized the significance of their imaging method: "Accessing the distal end of fiber devices and collecting sufficient training data are challenging in practical applications. The unique hollow organ or biological tissue environments impose additional difficulties in robust image transport."
"Yet, our Restore-CycleGAN-GALOF method requires only a small amount of training data and eliminates the need for pairing image features. In the small data regime, this method guarantees highly robust and strong generalizable full-color imaging. As a result, it is better suited to satisfy various practical biomedical applications."
"Our techniques are expected to lay the foundation for the next-generation fiber-optic imaging system. Our future research will focus on developing practical endoscopy systems and conducting related biomedical application tests. We aspire to advance medical diagnosis and fundamental biological research through our methodology," the scientists added.
More information: Xiaowen Hu et al, Unsupervised full-color cellular image reconstruction through disordered optical fiber, Light: Science & Applications (2023). DOI: 10.1038/s41377-023-01183-6
Journal information: Light: Science & Applications
Provided by Chinese Academy of Sciences