This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Chemical 'supercharger' solves molecular membrane mystery
Assemblies of tiny molecular proteins span the membranes that encapsulate our cells, directing cellular activities and regulating the transport of materials and information in and out.
In the fight against human disease, including various cancers and neurocognitive disorders, more than 60% of market drugs target these membrane proteins.
But a lack of technology has hampered scientists' efforts to capture key information about these proteins where they live, requiring the use of artificial chemical environments.
In an important breakthrough, scientists at the Yale Nanobiology Institute have decoded a chemical signal that allows them to capture these biological interactions directly from their natural habitat.
The finding, published in Nature Methods, opens up new avenues for understanding downstream cellular applications in human health.
"The traditional way to analyze these membrane proteins was to put them into an artificial environment, but that was a lot like studying a fish out of water," said Kallol Gupta, assistant professor of cell biology and lead author of the study.
Previous experiments relied on the "brute force" energy of conventional mass spectrometry techniques to remove proteins from their greasy membrane environments. But this damaged the proteins and their ability to bind with other molecules—including, crucially, those of potential therapeutic value.
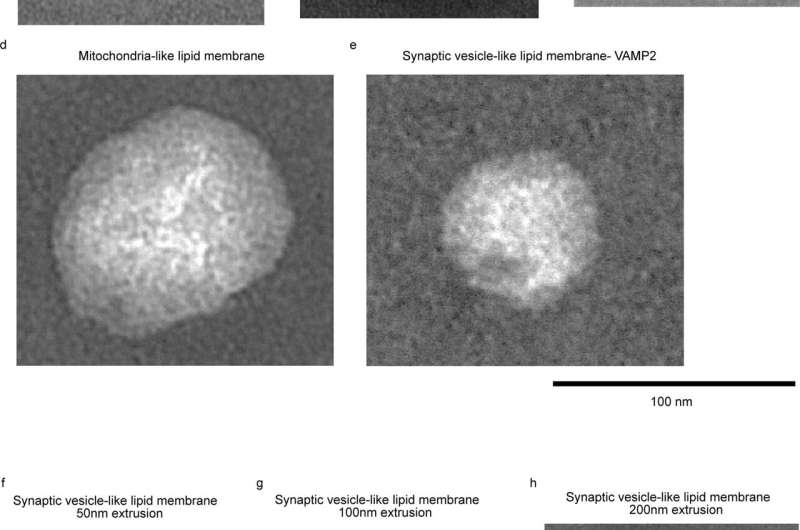
The team at Yale's West Campus identified a class of chemicals, called superchargers, that worked to gently destabilize the membrane while leaving the embedded proteins intact. They were then able to show how cell membranes regulate the speed of neurotransmitter release, a key step in central nervous system signaling.
This breakthrough technology opens the way for scientists to screen future therapeutics precisely and directly at the point where proteins encounter new drugs.
Aniruddha Panda, a postdoctoral researcher in the Gupta Lab, was the first author of the study, which included collaborators at the Rothman Lab, Yale Nanobiology Institute, the University of Oxford, Aarhus University, Texas Tech University and Sorbonne Université. Alongside an in-house mass spectrometer, the authors also utilized the West Campus Analytical Core and the Imaging Core in their experiments.
More information: Aniruddha Panda et al, Direct determination of oligomeric organization of integral membrane proteins and lipids from intact customizable bilayer, Nature Methods (2023). DOI: 10.1038/s41592-023-01864-5
Journal information: Nature Methods
Provided by Yale University