This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Anticancer drugs with fewer side-effects: Scientists decode the crystal structure of a key cell cycle protein
Anticancer drugs are pivotal to cancer treatment, but their toxicity may not always be limited to cancer cells, resulting in harmful side-effects. To develop anticancer therapies that have fewer adverse effects on patients, scientists are now focusing on molecules that are less toxic to cells.
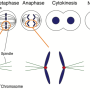
One such group of drugs is the "kinesin inhibitors." These inhibitors prevent cancer progression by explicitly targeting kinesin motor proteins, which are required for the division of cancer cells. Centromere-associated protein E (CENP-E), a member of the kinesin motor protein, is a promising target for inhibitor therapy, as it is essential for tumor cell replication. However, determining the structure of CENP-E is crucial to identify inhibitor molecules that can bind to CENP-E and arrest the function.
Interestingly, the binding of the energy molecule—adenosine triphosphate (ATP)—to the motor domain of CENP-E changes its structure or configuration. This also occurs when CENP-E binds to an inhibitor. So far, very few CENP-E inhibitors have been reported and none have been approved for clinical use. It is, therefore, important to acquire structural information on the CENP-E motor domain.
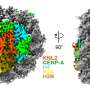
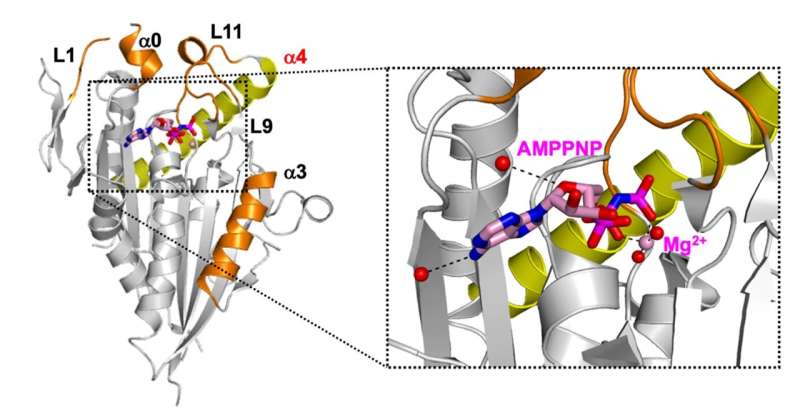
To this end, a research team from Tokyo University of Science (TUS) in Japan used X-ray crystallography to elucidate the crystal structure of the complex formed by the CENP-E motor domain and a kinesin inhibitor.
The study, which was led by Professor Hideshi Yokoyama from TUS, along with co-authors Ms. Asuka Shibuya from TUS, and Assistant Professor Naohisa Ogo, Associate Professor Jun-ichi Sawada, and Professor Akira Asai from the University of Shizuoka, was published in FEBS Letters.
"CENP-E selectively acts on dividing cells, making it a potential new target for anticancer drugs with fewer side-effects," says Dr. Yokoyama while discussing the motivation underlying this study.
First, the team expressed the CENP-E motor domain in bacterial cells, following which they purified and mixed it with adenylyl-imidodiphosphate (AMPPNP)—a non-hydrolyzable ATP analogue. The mix was crystallized to obtain exhaustive X-ray data. Using this data, the team obtained the structure of CENP-E motor domain-AMPPNP complex.
Next, they compared the structure with that of CENP-E-bound adenosine diphosphate (CENP-E-MgADP) as well as with other previously known kinesin motor protein-AMPPNP complexes. From these comparisons, the team speculated that the helix alpha 4 in the motor domain was likely to be responsible for the loose binding of CENP-E to microtubules, i.e., cell structures that are crucial to cell division.
"Compared to the α4 helices of other kinesins, the α4 of CENP-E binds slowly and with lesser strength to microtubules as compared to other kinesins, throughout the ATP hydrolysis cycle," adds Dr. Yokoyama.
The discovery of the crystal structure of the complex is expected to facilitate additional structure-activity relationship studies, which will bring scientists a step closer to developing anticancer drugs targeting CENP-E. The research team is optimistic about the future applications of their research and are confident that it will be possible to design drugs based on the methods employed in this study.
"The ultimate goal is to use the preparation and crystallization methods described in our study for future drug design studies that aim at developing anticancer drugs with fewer side-effects," concludes a hopeful Dr. Yokoyama.
More information: Asuka Shibuya et al, Crystal structure of the motor domain of centromere‐associated protein E in complex with a non‐hydrolysable ATP analogue, FEBS Letters (2023). DOI: 10.1002/1873-3468.14602
Journal information: FEBS Letters
Provided by Tokyo University of Science