This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Researchers reveal structural basis of plp2-mediated cytoskeletal protein folding by TRiC/CCT
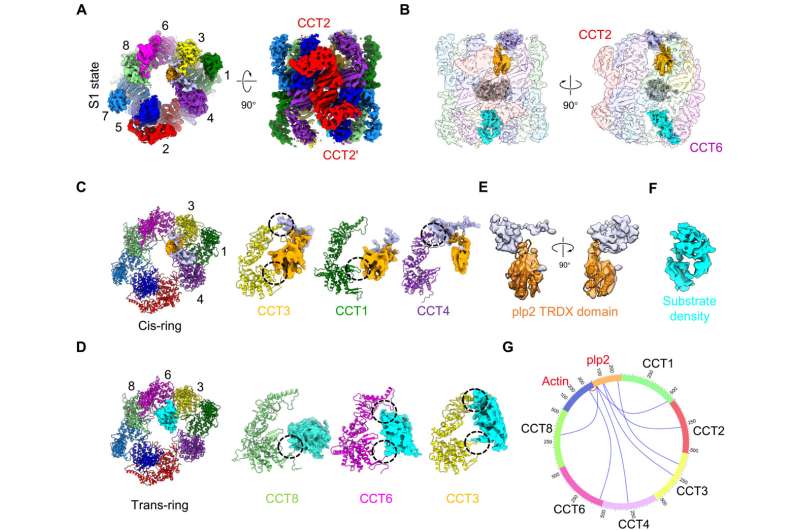
In a study published in Science Advances, Dr. Cong Yao's team from the Center for Excellence in Molecular Cell Science, Shanghai Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences, reported a complete picture of TRiC-assisted tubulin/actin folding along TRiC ATPase cycle under the coordination of co-chaperone plp2 through cryoelectron microscopy (cryo-EM) analysis.
The eukaryotic group II chaperonin TRiC/CCT assists the folding of ~10% of cytosolic proteins through ATP-driven conformational circle, including many key structural and regulatory proteins, such as the key cytoskeletal proteins tubulin and actin, the cell cycle regulator CDC20 and many proteins involved in oncogenesis. Thus, TRiC plays an essential role in maintaining cellular protein homeostasis. Dysfunction of TRiC is closely related to cancer and neurodegenerative diseases.
The major cytoskeletal proteins tubulin and actin are obligate substrates of TRiC. A remarkably complex cellular machinery consisting minimally of TRiC, cochaperone, and cofactors has evolved to facilitate their biogenesis. It has been shown that phosducin-like protein 2 (PhLP2) is essential for ciliogenesis and microtubule assembly, and the ciliary precursor tubulin needs to be folded by TRiC with assistance of PhLP2.
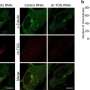
The researchers first determined an ensemble of cryo-EM structures of S. cerevisiae TRiC along its ATPase cycle, with simultaneously engaged plp2 and substrate actin or tubulin inside its chamber, one per ring, at the resolution of up to 3.05 Å, In the open S1/S2 states, plp2 and tubulin/actin engaged within opposite TRiC chambers, and the substrate density remains less well resolved, indicating that it might be in the initial stage of folding.
Intriguingly, the researchers captured an unprecedented TRiC-plp2-tubulin complex in the closed S3 state, engaged with a fully folded full-length β-tubulin which even loaded with a GTP since its "birth" from the TRiC chamber, and a plp2 occupying the opposite ring. This provides new clues for the biogenesis of tubulin and the assembly of a/b-tubulin heterodimers. Another closed S4 state revealed an actin in the intermediate folding state and a plp2.
Accompanying TRiC ring closure, plp2 translocation in one ring could coordinate substrate translocation on the CCT6 hemisphere of the opposite ring, facilitating substrate stabilization and folding. In addition, the co-chaperone plp2 engages within the cavity of TRiC regardless of the TRiC conformational state, but other co-chaperones of TRiC such as PFD and PhLP1 were observed only bound on the outer top of the open TRiC ring.
This study provides structural insights into the folding mechanism of the major cytoskeletal proteins tubulin/actin under the coordination of the complex biogenesis machinery TRiC and plp2, and could extend the understanding on the links between cytoskeletal proteostasis and related human diseases such as developmental and neurological disorders.
More information: Wenyu Han et al, Structural basis of plp2-mediated cytoskeletal protein folding by TRiC/CCT, Science Advances (2023). DOI: 10.1126/sciadv.ade1207
Journal information: Science Advances
Provided by Chinese Academy of Sciences