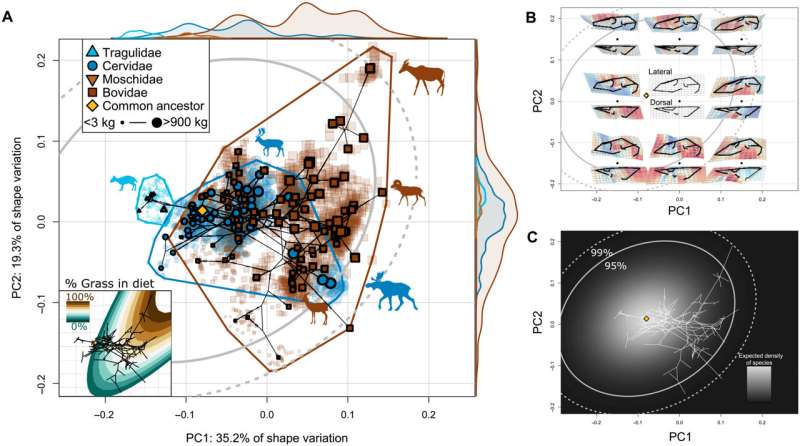
Evolutionary variation in the ruminant cranium is biased toward CREA. (A) Phylomorphospace of the species' mean cranial shapes, with individual specimens projected back into the space. Gray ellipses denote 95 and 99% confidence intervals of the probability distribution of species in morphospace given a Brownian motion model of evolution. Size of data point (species) corresponds to centroid size. Distributions shown on the edges of the plot represent the relative densities of each family. Inset: An ecological surface highlighting variation in percent grass in diet, where grazing species consume primarily grass and browsing species consume less grass. Note that variation in percent grass in diet is similar to variation in species' size. (B) Shape variation across the morphospace. Each black point represents a shape model at a given coordinate, with its lateral and dorsal views presented above and below, respectively. (C) Probability distribution of species in morphospace given our evolutionary rate matrix. Lighter areas denote more-likely areas a species will exist in morphospace given a Brownian motion model of evolution. An interactive version of this morphospace (and others) is available for nonmobile devices (https://danielrhoda.shinyapps.io/Ruminant_Dashboard/). Credit: Science Advances, doi: 10.1126/sciadv.ade8929
Evolutionary biologists aim to form fundamental connections between microevolutionary processes and macroevolutionary patterns based on comparative datasets of population-level variation. In a new report on Science Advances, Daniel R. Rhoda and a team of scientists in evolutionary biology at the University of Chicago and the Jackson Laboratory in the U.S., analyzed a previously published dataset of ruminant (mammalian) crania.
The results were biased via highly conserved craniofacial evolutionary allometry (CREA), where larger species demonstrated proportionally longer faces. The outcomes highlighted the feature as an evolutionary line of least resistance, facilitating morphological diversification aligned with the browser-grazer spectrum. The results show how constraints at the population level can produce highly directional patterns of phenotypic evolution at the macroevolutionary scale. The work sheds light on exploring the role of craniofacial evolutionary allometry across mammalian clades.
Craniofacial evolutionary allometry (CEA)
Natural selection affects phenotypic variation in a population, where the development of the population responds to selection. The direction of the greatest amount of variation is known as the line of least resistance (LLR) and represents the potential direction of greatest evolutionary change. If the selection is aligned with the line of least resistance, biologists expect populations to evolve in a direct path towards an adaptive peak. However, if the selection is oriented elsewhere, the response to selection will be realigned towards the line of least resistance. As a result of this, interactions between the adaptive landscape and limits of variation within a species at the population level determine the path of phenotypic evolution.
Evolutionary biologists seek to explain global patterns of biodiversity as a fundamental goal in biological research to uncover the microevolutionary mechanisms underlying macroevolutionary patterns. They expect these patterns to influence population-level constraints on macroevolution. In this work, Rhoda et al. present strong evidence to the influence of craniofacial evolutionary allometry (i.e. characteristics of living creatures that change with size) on micro- and macro-evolution, which can be explored alongside the line of least resistance to facilitate morphological diversity.
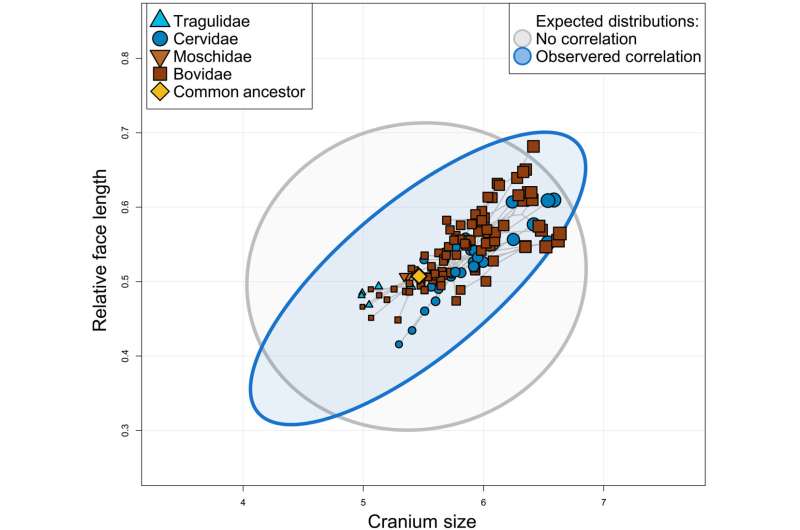
Larger ruminant species have proportionally longer faces, as predicted by CREA (Craniofacial evolutionary allometry). The gray ellipse represents the 95% confidence interval of the expected distribution of species if there was no allometry of face length (given the observed evolutionary rates), and the blue ellipse represents the expected distribution of species given the observed evolutionary rate matrix. Note that certain "extreme" trait combinations (e.g., very large and long-faced species) are only accessible under correlated evolution, whereas other combinations (e.g., very small, long-faced species) are only accessible under hypothetical uncorrelated evolution. Credit: Science Advances, doi: 10.1126/sciadv.ade8929
The highly conserved pattern of the mammalian skull
The mammalian skull maintains a highly conserved pattern of ontogenetic and evolutionary allometry, where larger species have proportionally longer faces as an example of craniofacial evolutionary allometry. While allometry is a limit on phenotypic evolution, it presents an opportunity for extreme phenotypes to arise without developmental novelty.
For instance, larger, short-faced mammals are improbable while the long-faced, out-of-reach phenotypes arise due to this allometry. Craniofacial evolutionary allometry therefore appeared as an evolutionary line of least resistance, simply indicating that larger mammals had longer faces due to size-related limits on skull formation. The paleontologists noted a strong relationship between the size and skull shape between species, where larger species represented longer faces, and the slope of evolutionary allometry significantly varied between the subfamilies.
Life through time, a visual timeline - artistic representation of the Neogene period - Neogene scene. An artistic reconstruction of a Wooly Mammoth behind an apple tree , early horses in the background, a Terror Bird, a flying insect, a Big Brown Bat, a crocodile in the pond, a Saber-tooth Cat on the shore, and Ground Sloths in the forest. Credit: Natural History Museum, Arcata California https://natmus.humboldt.edu/exhibits/life-through-time/visual-timeline/neogene-period
Cranial shapes between species
Example work showed the cranial shapes of dik-diks to be unlike other small ruminants due to greater nasal retraction. Yet, all small ruminants possessed shorter faces than their more massive, long-faced relatives. Using phylogenetic principal component analysis, the team showed clear size trends dominating the major axes of variation to further strengthen the results. Rhoda et al. recreated the interspecific morphospace between clades to better understand diverse evolutionary patterns while exploring the craniofacial evolutionary allometry.
The team noted and corroborated the morphospace similarities in the analysis to be due to allometry—the primary influence on bovid skull diversification. The researchers noted a clear size trend for evolutionary variation in the bovid cranium, where the large, grazing tribe Alcephini and the small browsing tribe Neotragini diversified more than expected relative to their observed evolutionary rate. The work was in agreement with the macroevolutionary patterns of diversification within bovids and cervids that appeared to diverge further away from their ancestor. The data represented the evolutionary lability of the ruminant diet through the Neogene period, while exemplifying the declining diversity of perissodactlys throughout the Cenozoic era.
The adaptive significance of CREA across species and the dynamics of microevolution
Rhoda et al. examined the shape variation associated with the browser-grazer continuum and were unclear if the exceptionally long-faced crania of large grazing species such as Alcelaphini resulted from (1) direct selection of crania better adapted for grazing, which coincidentally happened to be long-faced, or (2) were agnostic to selection for foraging ecology. In the latter scenario, their assumption was that longer faces passively evolved during increases in body sizes associated with grazing, which seemed unlikely due to the intimate connection between food ingestion and processing.
In either probable scenario, the foraging ecology provided adaptive value for evolutionary changes in size and influenced the ruminant cranial morphology to diversify. Rhoda et al. examined if the craniofacial evolutionary allometry (CREA) is an evolutionary line of least resistance in ruminant artiodactyls and analyzed its influence on morphological diversity. They directly measured the axis of variation associated with a hypothetical CREA to interrogate its relationship with evolutionary variation at the population level and conducted phylogenetic generalized least-square regressions to measure associations between morphological divergence and the angle of divergence relative to CREA.
-
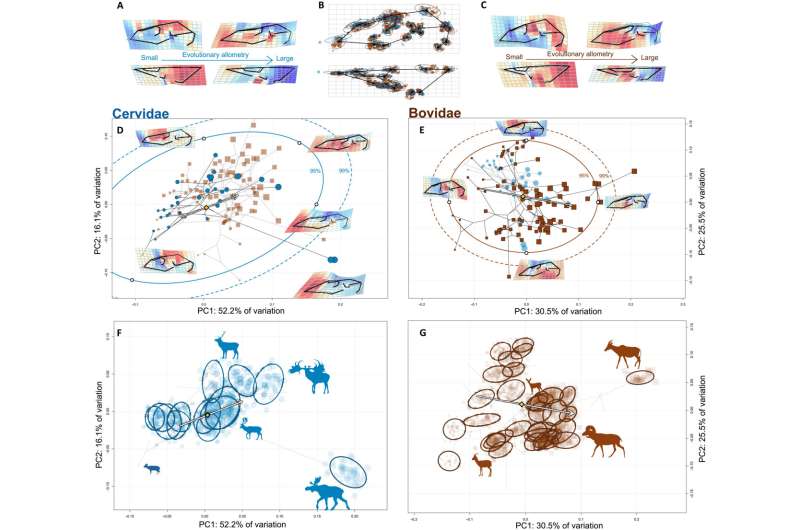
CREA governs phenotypic variation within both Cervidae and Bovidae. (A and C) Evolutionary allometric trajectories of the cervid (A) and bovid (C) cranium. (B) Mean shape of the dataset, with the Procrustes superimposed landmark scatters colored by family and scaled according to species' centroid size. (D and E) Phylomorphospaces of Cervidae and Bovidae, with confidence intervals of the probability distribution of species in morphospace. Arrows denote allometric trajectories, as in (F) and (G). (F and G) Family-specific morphospaces with individual specimens projected into them and ellipses for each species in the intraspecific dataset. Credit: Science Advances, doi: 10.1126/sciadv.ade8929
-
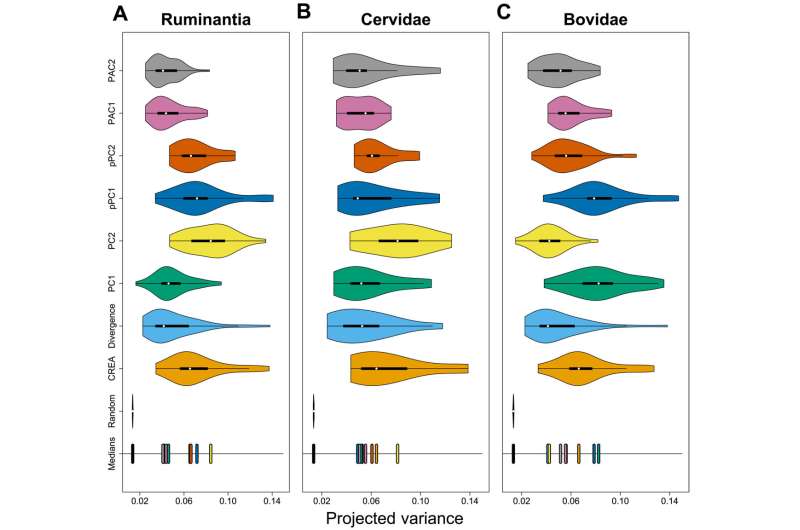
CREA explains as much intraspecific variation as the major axes of interspecific variation. Distributions of species' projected variance values for different directions of evolutionary variation for all of Ruminantia (A), Bovidae (B), and Cervidae (C). Axes of evolutionary variation for Cervidae and Bovidae are calculated using only members of the respective clade. The positions of medians of the distributions are at the bottom of each plot Credit: Science Advances, doi: 10.1126/sciadv.ade8929
Outlook
In this way, Daniel R. Rhoda and colleagues examined the microevolutionary processes underlying macroevolutionary patterns during the diversification of the mammalian (ruminant) skull. They explored the craniofacial evolutionary allometry (CREA) as an aspect of the line of least resistance (LLR), so when they generated matrices of species during the work, the outcomes closely aligned to CREA; indicating that the species had diverged farther from their ancestors than those less-aligned.
The work examined intrinsic and extrinsic factors underlying the differences among clades and their patterns of diversification relative to bovids and cervids in the browser-grazer spectrum. And highlighted the influence of intrinsic factors on the diversification of skull morphology of ruminants. The outcomes broadly demonstrated the influence of intrinsic limits on macroevolutionary patterns, allowing the scientists to envision further research on the role of CREA in other mammalian clades.
More information: Daniel P. Rhoda et al, Diversification of the ruminant skull along an evolutionary line of least resistance, Science Advances (2023). DOI: 10.1126/sciadv.ade8929
Ryan N. Felice et al, Developmental origins of mosaic evolution in the avian cranium, Proceedings of the National Academy of Sciences (2017). DOI: 10.1073/pnas.1716437115
Journal information: Proceedings of the National Academy of Sciences , Science Advances
© 2023 Science X Network