This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Researchers report novel cell size regulation mechanism in cyanobacteria
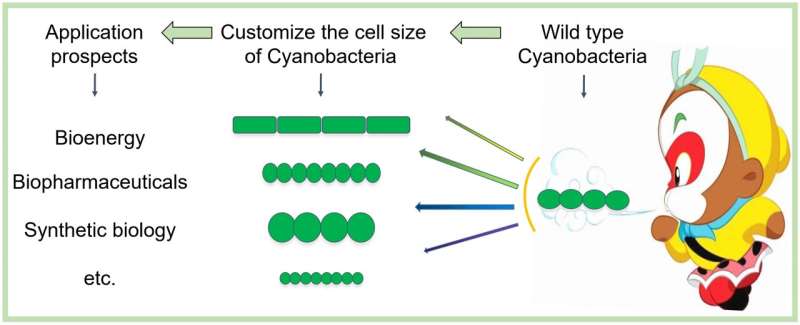
Cyanobacteria are the earliest known oxygenic photosynthetic organisms on Earth, and they played decisive roles in the evolution of the environment and the life on our planet. Cell morphology and cell size of different cyanobacteria species vary widely, but each species has inheritable and distinct cell morphology and cell size that are stably maintained through the generations. The underlying mechanisms of such a homeostasis have been unknown.
In a study published in Proceedings of the National Academy of Sciences, the research group led by Prof. Zhang Chengcai from the Institute of Hydrobiology (IHB) of the Chinese Academy of Sciences reported a new cell size regulation mechanism which represents an important advance in understanding cell morphology and cell size controlling of cyanobacteria.
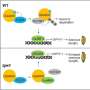
Previous studies by Prof. Zhang's group have showed that the second messenger c-di-GMP may be an intracellular proxy for cell size control in the filamentous cyanobacterium Anabaena PCC 7120. However, it was unclear how this chemical signal is perceived in cells. In addition, all c-di-GMP receptors known in other organisms are not conserved in cyanobacteria, posing a challenge to understand the physiological function of c-di-GMP in cyanobacteria.
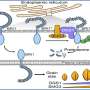
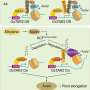
In this study, the researchers identified and characterized the first c-di-GMP receptor, CdgR, from the cyanobacterium Anabaena. Crystal structural analysis and genetic studies showed that CdgR binds c-di-GMP at the dimer interface, and this binding is required for the control of cell size in a c-di-GMP-dependent manner. Combining biochemistry analysis with genetic analysis, the researchers discovered that CdgR functions by interacting with a global transcription factor DevH, and that this interaction was inhibited by c-di-GMP.
"This study provides a novel mechanism of cell size regulation. Some mutants associated with genes of this pathway produce a lethal phenotype, indicating that this signaling pathway plays a critical function in this cyanobacterium. The exploration of the cell size and cell morphology control is critical for customizing and constructing cyanobacterial cells with particular features adapted to biotechnological applications using synthetic biology approach," Prof. Zhang said.
CdgR is highly conserved in cyanobacteria. Therefore, this study lays the groundwork for the understanding of the roles of c-di-GMP signaling in these organisms, including important physiological processes such as photoregulation, biofilm formation, cell motility, and cyanobacteria bloom formation.
Prof. Zhang's group has been devoted to basic research on prokaryotic cell morphogenesis and synthetic biology research related to the cyanobacterial bloom control. Former findings on the coordination mechanism of cell division and development by this group have been published in PNAS and PNAS Nexus.
More information: Xiaoli Zeng et al, A c-di-GMP binding effector controls cell size in a cyanobacterium, Proceedings of the National Academy of Sciences (2023). DOI: 10.1073/pnas.2221874120
Journal information: Proceedings of the National Academy of Sciences , PNAS Nexus
Provided by Chinese Academy of Sciences