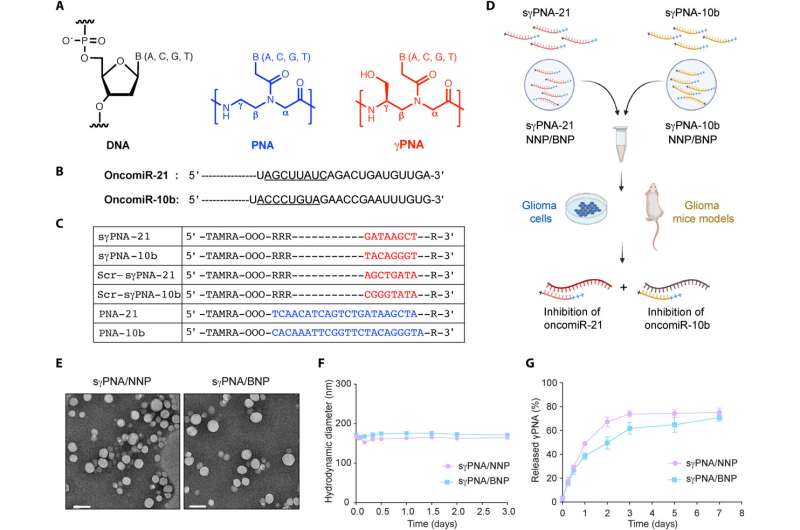
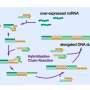
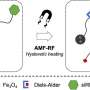
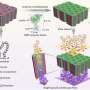
Design of PNAs and characterization of NPs. (A) Chemical structure of DNA, PNA, and serine-γ-PNA. (B) The sequence of oncomiR-21 and oncomiR-10b, where the underlined segment is the seed region. (C) PNAs used in the study. sγPNA-21 and sγPNA-10b are serine-γPNAs designed to bind the seed region of oncomiR-21 and oncomiR-10b, respectively. Three arginine (RRR) residues are appended to the N terminus and one arginine (R) on the C terminus. Scr-sγPNA-21 and Scr-sγPNA-10b are scrambled versions of sγPNA-21 and sγPNA-10b, respectively. PNA-21 and PNA-10b are regular PNAs designed to bind full-length oncomiR-21 and oncomiR-10b, respectively. PNAs were conjugated with TAMRA, a fluorescent dye for imaging. OOO represents 8-amino-2,6,10-trioxaoctanoic acid residues (Mini-PEG). (D) Graphical representation of sγPNA encapsulation in NPs (NNP or BNP) and treatment strategy for the simultaneous inhibition of oncomiRs 21 and 10b. (E) TEM images of sγPNA/NNP and sγPNA/BNP. Scale bars, 100 nm. (F) Size stability of sγPNA/NNP and sγPNA/BNP in aCSF. (G) The amount of sγPNA (sγPNA-21 and sγPNA-10b) released from NPs over time during incubation in buffered saline was determined and quantified as a percentage of the amount loaded. The data are shown as means ± SD (n = 3). Credit: Science Advances (2023). DOI: 10.1126/sciadv.abq7459
A team of researchers, including members from the University of Connecticut, has developed a nanoparticle-based treatment that targets multiple culprits in glioblastoma, a particularly aggressive and deadly form of brain cancer.
The results, a collaboration between UConn and Yale University, were published today in Science Advances.
The new treatment uses bioadhesive nanoparticles that adhere to the site of the tumor and then slowly release the synthesized peptide nucleic acids that they're carrying. These peptide nucleic acids target certain microRNAs—that is, short strands of RNA that play a role in gene expression. Specifically, they're directed at a type of overexpressed microRNA known as "oncomiRs" that lead to the proliferation of cancer cells and growth of the tumor. When the peptide nucleic acids attach to the oncomiRs, they stop their tumor-promoting activity.
The laboratories of professors Raman Bahal of the UConn School of Pharmacy and Mark Saltzman of Yale collaborated on the treatment system. Unlike similar efforts, which target only one oncomiR at a time, this treatment targets two, making its effect on cancer cells stronger. The test mice who received the treatment lived for a significantly longer time than the control mice.
"It can knock down both targets at the same time, which turns out to have a remarkably more powerful result, as we saw with the increased survival results," says Saltzman, a professor of biomedical engineering, chemical and environmental engineering, and physiology. "These results are the best I've ever seen in this sort of aggressive brain tumor."
One challenge in developing the treatment was designing the anti-cancer agents, known as antimiRs, so that two different ones could fit in a single nanoparticle.
"We synthesized all these compounds and came up with the idea that you don't have to target one oncomiR at a time," says Bahal, an associate professor of pharmaceutics. "Now we can think about multiple oncomiR targets."
For this work, the researchers targeted the oncomiRs known as miR-10b and miR-21, which are both very common in glioblastoma. Future treatments, though, can be easily tailored for specific patients. For instance, if a biopsy of a patient's tumor produces a profile showing the proliferation of different oncomiRs, the treatment could be appropriately altered.
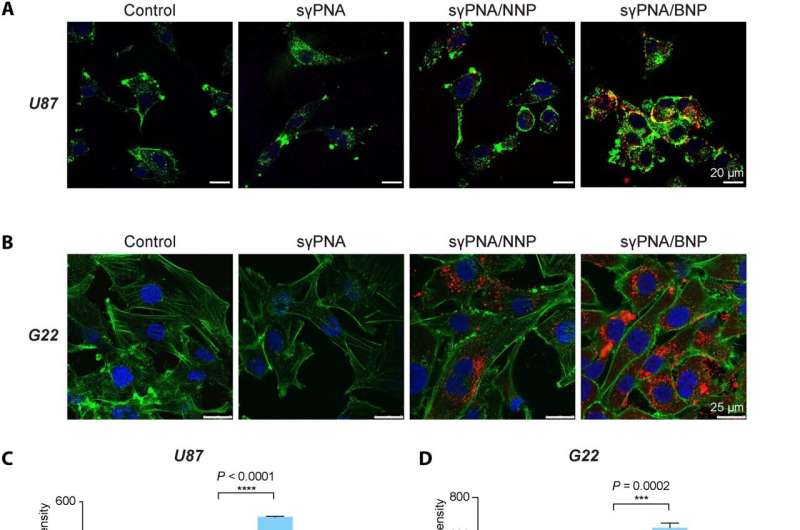
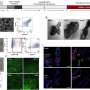
sγPNA-mediated simultaneous knockdown of miR-21 and miR-10b in multiple glioma cells. (A) Confocal microscopic image of U87 cells treated with free sγPNA or sγPNA-loaded NPs. Scale bars, 20 μm. (B) Confocal microscopic images of G22 cells (patient-derived GBM cells) treated with free sγPNA or sγPNA-loaded NPs. Scale bars, 25 μm. PNAs were conjugated with TAMRA (red); F-actin was labeled with phalloidin (green), and the nucleus was stained with Hoechst (blue). (C) Cellular uptake in U87 cells analyzed by flow cytometry. Data are expressed as means ± SD (n = 3). (D) Quantification of cellular uptake of sγPNA in G22 cells via flow cytometry analysis. Data are expressed as means ± SD (n = 3). (E) Expression analysis of miR-10b and miR-21 levels in U87 cells after treatment with Scr-sγPNA/NNP, Scr-sγPNA/BNP, sγPNA/NNP, sγPNA/BNP, and free sγPNA. (F) The levels of miR-10b and miR-21 in G22 cells treated in vitro with sγPNAs (sγPNA-10b + sγPNA-21) and BNPs containing sγPNAs and scr-sγPNAs. sγPNA/BNPs are a physical mixture of sγPNA-21 BNP and sγPNA-10b BNP. sγPNA/NNPs are a physical mixture of sγPNA-21 NNP and sγPNA-10b NNP. NNP indicates PLA-HPG NP, and BNP indicates PLA-HPG-CHO NP. Credit: Science Advances (2023). DOI: 10.1126/sciadv.abq7459
Saltzman calls the treatment "a marriage of two technologies."
"One is the bioadhesive nanoparticle technology, which we had developed earlier, and marrying it to this peptide nucleic acid technology that Raman has perfected," he says.
Because the treatment is localized to the tumor site, Bahal noted that both the synthesized nucleic acids and the nanoparticles that deliver them to the tumor site are non-toxic. Also critical to the treatment's success is that the particles and the agent it releases remain at the tumor site for about 40 days. Conventional site-specific treatments tend to wash away fairly quickly.
"These are high-binding molecules that are scalable and effective simultaneously," says Bahal. "It's targeted and stays there. Traditional molecules have had many challenges in terms of toxicity."
Ideally, the delivery system would be applied as part of a larger treatment regimen.
"We designed it to be an add-on to what physicians do already," Saltzman says. "They would do a surgery, then they infuse our nanoparticles, and then they give chemotherapy and/or radiation in the way that they normally do. We're expecting that this would lead to a better result because the nanoparticle/anti-microRNA is sensitizing the cells to the chemotherapy and the radiation therapy."
The study's other authors include Shipra Malik, a former graduate student in Bahal's lab; and Vijender Singh, Associate Director of Computational Biology Core at UConn.
"This study opens new avenues for RNA-targeted technology in developing personalized therapy for brain cancer," says Malik.
More information: Yazhe Wang et al, Anti-seed PNAs targeting multiple oncomiRs for brain tumor therapy, Science Advances (2023). DOI: 10.1126/sciadv.abq7459
Journal information: Science Advances
Provided by University of Connecticut