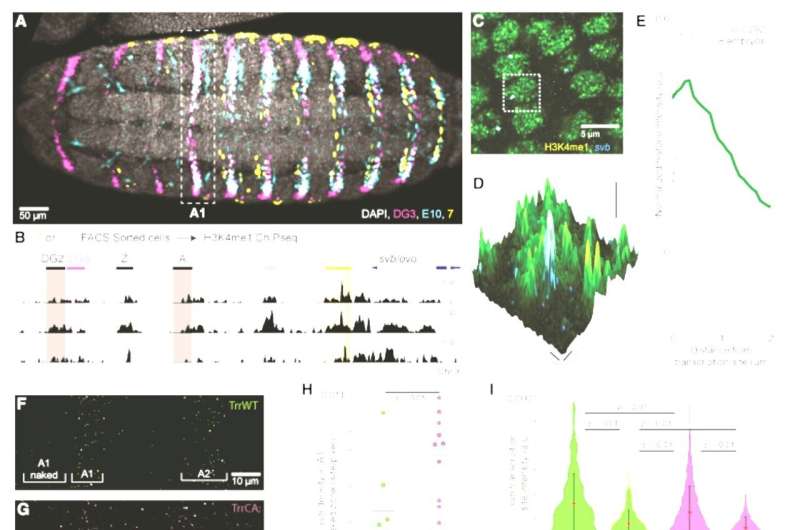
H3K4me1 at the svb locus supports transcriptional robustness and microenvironment integrity(A) Ventral view of a stage 15 Drosophila melanogaster embryo from the fly line used for the ChIP-seq experiment stained for the products of the reporter genes driven by the svb enhancers DG3, E10, and 7. The white dotted box is the first abdominal (A1) segment.(B) ChIP-seq using cells from stage 15 Drosophila melanogaster embryos sorted by reporter gene activity. The panel shows H3K4me1 enrichment at the svb/ovo locus, in cells with an active 7 enhancer (orange), an active E10 enhancer (cyan), or from the entire embryo (“All,” gray). The darker shade in the 7 enhancer highlights the “H” region, which encompasses most of its reported regulatory activity, where the enrichment of H3K4me1 is most conspicuous.(C) Confocal imaging experiments in stage 15 w1118 embryos show that svb transcription sites are in regions enriched for H3K4me1. The dashed box highlights a single nucleus of the embryonic epidermis.(D) Zoomed-in view of a single nucleus (dotted box in C) with the height indicating the intensity of the H3K4me1 signal. svb RNA is in cyan.(E) Normalized average H3K4me1 intensity over 292 transcription sites in 8 embryos. The shaded region is the variance.(F and G) svb transcription sites at 25°C on the ventral side of the first two abdominal (A1 and A2) segments of stage 15 embryos in both trr1 lines.(H) Transcription site density in front of the A1 ventral band (“A1 naked”). Number of embryos: 7 (TrrWT) and 10 (TrrCA). The center line is the mean. The boxed region is one SD, and the tails are two SDs (95%).(I) Intensity of svb transcription sites at different temperatures. The red dot is the mean, and the bar is two SDs.(J) Confocal microscopy image of active svb transcription sites and Ubx distribution in the A1 segment of a stage 15 TrrWT embryo.(K and L) Confocal imaging experiments in stage 15 embryos show that H3K4me1 hypomethylation impairs Ubx enrichment at svb transcription sites. Right panels: zoomed-in view of the dotted boxes with the height indicating the intensity of the Ubx signal.(M and N) Intensity of the Ubx signal in svb transcription sites measured in the ventral (M) or in the lateral region (N). The red dot is the mean, and the bar is two SDs. Number of embryos: TrrWT = 5 embryos, TrrCA = 8 embryos. Number of analyzed transcription sites in the ventral region (M): TrrWT n = 69 and TrrCA = 139 and in the lateral region (N): TrrWT n = 38 and TrrCA n = 45.All p values in the figure are from two-tailed Student’s t tests. n.s., not significant. Credit: Cell Reports (2022). DOI: 10.1016/j.celrep.2022.111832
Most of his career, Justin Crocker, EMBL Heidelberg Group Leader, has been working at the interface of development and evolution. In two new studies led by Crocker, scientists have shown how using non-standard laboratory conditions and synthetic biology approaches can help us understand fundamental mechanisms that regulate the development and evolution of phenotypes.
In this, they are part of the growing field of phenomics—the systematic study of an organism's traits and how they vary and change during development as well as in response to the environment. Phenotypic evolution becomes particularly interesting in light of global concerns such as climate change, where many animals are under pressure to adapt quickly to fast-changing environments.
The significance of studying phenotypes
Phenotypes are the observable characteristics of an organism—features such as behavior, appearance, metabolism, gene expression patterns, etc. They result from interactions between the genotype—the information contained in DNA, and the environment. The phenotypes any organism exhibits often depend on precise decisions regarding which genes are expressed where and when.
In the two publications, the Crocker group and their collaborators provide novel insights into some of the key processes that determine the robustness of phenotypes and the appearance of new phenotypes during development.
This knowledge can help researchers better understand how diversity emerges during evolution in animals, and perhaps even predict ecological and environmental patterns of change in the phenotypes of wild animal populations.
Studying phenotypes in laboratories vs. 'the wild'
Biologists often study organisms under well-standardized laboratory conditions to ensure rigor and reproducibility. However, this also increases the risk of missing effects that only become apparent outside of these narrow ranges of conditions.
Using fruit-fly embryos and a variety of other model systems, Crocker and his team have been demonstrating the importance of moving beyond standardized laboratory conditions and challenging established assumptions when it comes to understanding the development and evolution of phenotypes.
The team used food sources incorporating fruit grown in and around the EMBL Heidelberg campus in their experiments that bridge natural and lab environments to understand the evolution of phenotypes.
In their experiments, they found that the loss of a certain epigenetic mark, the H3K4 monomethylation, led to changes in behavior, gene expression, metabolism, and even rates of offspring production.
"This epigenetic mark is present throughout the genome, but its deletion seems to have little to no impact on gene expression, which led scientists to hypothesize that it doesn't play a major role in normal development and function," said first co-author Albert Tsai, team leader at the Centre de Recherche en Biologie cellulaire de Montpellier (CRBM), former postdoc in the Crocker lab.
H3K4 monomethylation is found ubiquitously in almost every cell's nucleus. "That led us to question why there is such an evolutionary drive to create these marks if it's actually doing nothing," said Tsai. The Crocker group was sure that they were missing something.
The scientists observed that the loss of H3K4 monomethylation led to changes, especially when the fruit-flies were fed on natural food sources, including fruit collected in and around the EMBL campus. In the absence of this mark, certain traits became sensitive to environmental conditions and to different genetic backgrounds. When exposed to high temperatures or when certain background genes were mutated, the organisms likewise responded differently.
"It challenges the current paradigm of standardizing experiments as much as possible to focus on very specific conditions," said Tsai. "We need to come up with controlled ways of bringing more natural environments into the lab."
Synthetic biology and the study of phenotypes
In a second study, Crocker and his team questioned how new phenotypes emerge in the first place. This is a central question in evolutionary biology—for organisms to accumulate small changes that would be selected by the environment, there must be a way to continuously, quickly, and easily introduce variation in phenotypes.
While our genomes often accumulate small changes—called mutations—over time, these don't always result in changes in phenotypes, or observable traits.
The team began by studying the expression of various genes in fruit fly embryos with point mutations—single-nucleotide DNA changes—in enhancer regions of the genome. "What we quickly started to appreciate was that while gene expression levels changed in these mutants, it always remained within the same regions," said Rafael Galupa, first author of the paper and former postdoc in the Crocker lab.
In other words, if a gene is usually active in the gut, for example, its expression levels increased or decreased as a result of the mutations, but did not shift to a different tissue, e.g. the muscle. "So we started wondering, what does it take to get expression elsewhere?" said Galupa, who is currently on his way to establishing his independent lab in Centre de Biologie Intégrative, Toulouse (France). "Ultimately in the course of evolution, how do you get new functions?"
Next, the team introduced completely random sequences into the genome instead. In a natural context, random DNA sequences may arise in the genome due to viruses, or transposons—mobile genetic elements that actively move between different parts of the genome.
To the researchers' surprise, with their synthetic DNA approach, they found that random sequences easily drove gene expression, and in all parts of the embryo.
"We have been talking about doing this for a long time, and everybody thought it was a bit crazy. Then we just went ahead and did it," said Crocker. "In the field we often think about how expression is generated, how to activate genes, etc. This study makes us think that if any random sequence can drive expression, and we have a genome with millions of sequences—maybe the question is not so much how do you generate expression but how do you repress or control it."
In future studies, the Crocker lab will continue to look deeply into the mechanisms that connect genotype and environment to phenotypes.
The studies appeared Cell Reports and Developmental Cell.
More information: Lautaro Gandara et al, Developmental phenomics suggests that H3K4 monomethylation confers multi-level phenotypic robustness, Cell Reports (2022). DOI: 10.1016/j.celrep.2022.111832
Rafael Galupa et al, Enhancer architecture and chromatin accessibility constrain phenotypic space during Drosophila development, Developmental Cell (2023). DOI: 10.1016/j.devcel.2022.12.003
Journal information: Developmental Cell , Cell Reports
Provided by European Molecular Biology Laboratory