Stage-specific transcriptional divergence regulation model in wheat embryogenesis. Credit: IGDB
Embryogenesis is one of the most fundamental and remarkable processes in both animals and plants. It's amazing that after fertilization, a single maternal egg cell can develop into an organism with a multilayered body plan only in just a few weeks. Cell fate transition is largely determined by the expression of the associated genes and the epigenetic state, which can influence gene expression. There are conserved and distinct features in the cellular process for embryogenesis in animals and plants. Although many studies have been published on animal embryogenesis, gene expression and epigenetic changes during plant embryogenesis are still elusive.
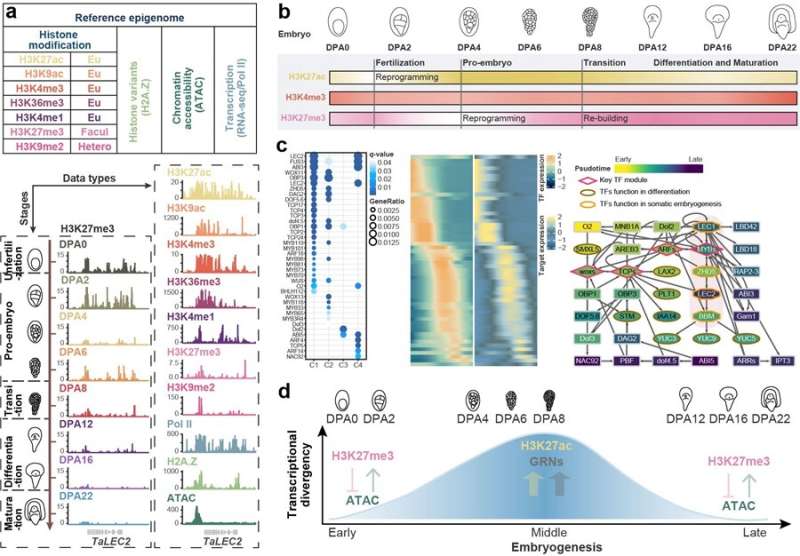
Prof. Xiao Jun's team from the Institute of Genetics and Developmental Biology (IGDB) of the Chinese Academy of Sciences (CAS) focused on the allohexaploid wheat and constructed a reference epigenome of wheat embryogenesis. The reference epigenome included seven histone modifications (H3K4me1, H3K4me3, H3K9ac, H3K9me2, H3K27ac, H3K27me3, and H3K36me3), occupancy of histone variant H2A.Z and RNA polymerase II, and chromatin accessibility, and transcriptomes.
The reference epigenome profiled the transcription and chromatin state dynamics during embryonic development, and provided clues to the regulatory mechanisms underlying cooperation and conflict among sub-genomes within a hexaploid.
According to their latest research, there was a huge transcription and chromatin state change following fertilization in wheat embryo. The histone modification H3K27ac (active gene expression) was decreased at 2 days post anthesis (DPA2) and marked mainly floral genes. Resetting of H3K27ac can silence these genes and detach the zygotic from the egg cell. H3K27me3 (repress gene expression) was reduced at DPA4, and marked mainly stem cell niche-related genes. The resetting of H3K27me3 may activate these genes and facilitate cell division. This result suggests that the reduction of H3K27ac and H3K27me3 may contribute to zygotic activation (ZGA) in wheat.
Interestingly, this epigenetic dynamic pattern was different from that in animals, although the cellular process of maternal to zygotic transition is similar between animals and plants.
During mid-embryogenesis in wheat, the chromatin accessibility and H3K27ac modification were increased, creating a permissive chromatin environment. This chromatin state was conducive to the binding of transcription factors to the cis-regulatory regions of genes.
Based on the cis- and trans-regulation features, the researchers constructed the gene regulatory networks for embryo pattern, which could facilitate the gene function dissection. In late-embryogenesis, the chromatin was condensed again, with an increase in H3K27me3. The condensed chromatin was also found around the differentiation-related genes, which kept the genes silent. This may be the reason why plants cannot go through deeper organogenesis like animals can.
Taken together, the chromatin state was condensed in early- and late-embryogenesis, but accessible in mid-embryogenesis in wheat. This was correlated with the similar-different-similar expression pattern from the sub-genome comparison.
In addition, different epigenetic modifications and transposable elements insertion at the three sub-genomes may also affect the bias expression of homoeologs. As a plant with a large genome, the distal regulation in wheat is more important and complex than in plants with smaller genome sizes such as Arabidopsis and rice.
This work provides an unprecedented epigenomic resource for embryogenesis research in wheat, which can facilitate the functional study of key genes during embryogenesis, especially in ZGA.
This study was published in Genome Biology, titled "Dynamic chromatin regulatory programs during embryogenesis of hexaploid wheat."
More information: Long Zhao et al, Dynamic chromatin regulatory programs during embryogenesis of hexaploid wheat, Genome Biology (2023). DOI: 10.1186/s13059-022-02844-2
Journal information: Genome Biology
Provided by Chinese Academy of Sciences