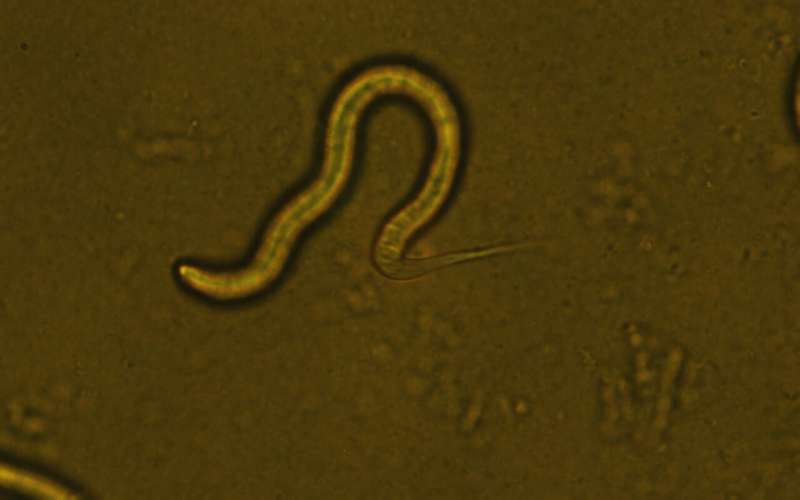
A research team from HKUST has solved the puzzle of C. elegans pri-miRNA processing. Credit: HKUST
The study of microRNAs (miRNAs), small RNAs that play important roles in gene regulation in animals and humans alike, have long been a topic of research interest. How these miRNAs control and regulate gene expression is believed to hold the key to the development of effective treatment strategies for conditions such as cancer, which is a result of cell mutations.
While miRNAs and their biogenesis in humans attracts the most interest from scientists, the study of Microprocessor, a protein complex that initiates miRNA biogenesis, is rarely reported. Recently, a group of scientists at the Hong Kong University of Science and Technology (HKUST) mapped out the fundamental mechanisms of C.elegans Microprocessor (cMP), paving the way for future studies into an area that could provide a wider perspective on how miRNAs function as a whole across living beings.
Their research was published in the open-access journal, Nucleic Acids Research.
"The molecular mechanism of C. elegans Microprocessor (cMP) has remained elusive since its discovery 18 years ago," said Prof. Tuan Anh Nguyen, principal investigator of the paper and Assistant Professor, Division of Life Science, HKUST. "Surely, the interest of many has been drawn to the study of miRNA in human beings for good reasons. But the lack of information and fundamental understanding in this complex in C. elegans has driven us to dive in."
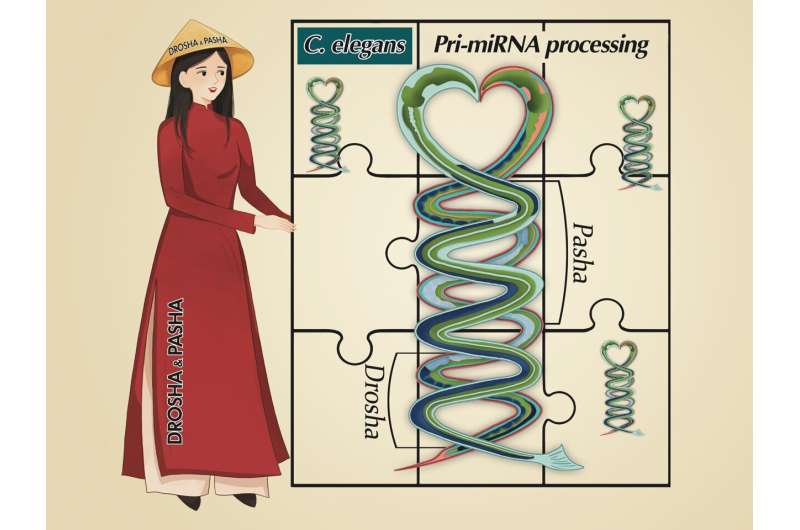
The pri-miRNA structure is illustrated by two C. elegans. Drosha and Pasha are coordinated to process pri-miRNAs. Credit: HKUST
Prof. Nguyen and his team investigated the molecular mechanism of cMP by conducting high-throughput pri-miRNA cleavage assays. In the process, they were able to reveal cMP's distinctive molecular mechanism.
"We demonstrated that cMP consists of two subunits, cDrosha and Pasha, and each has its own ability to measure the stem lengths of C. elegans pri-miRNAs ," he said. "These two subunits can determine the cleavage sites of the complex using their distinct measuring methods, but more importantly, the mechanism we revealed is different from human MP (hMP) in many aspects. For example, human DROSHA measures only 13 bp and determines the cleavage sites of hMP, whereas DGCR8 (a Pasha orthologue) does not appear to have the ability to either measure or determine the cleavage sites at all."
With the mechanisms of cMP now apparent, Prof. Nguyen plans to further investigate the structure of cMP/pri-miRNA.
"We now know that dsRBDs and linkers of Pasha are necessary for the 25-bp upper stem measurement, but as far as fully understanding the structural basis of these substrate numbers, we are just getting started, and we think many in the field will find it worthwhile to pursue further," said Prof. Nguyen.
More information: Thuy Linh Nguyen et al, Dissection of the Caenorhabditis elegans Microprocessor, Nucleic Acids Research (2023). DOI: 10.1093/nar/gkac1170
Journal information: Nucleic Acids Research
Provided by Hong Kong University of Science and Technology