This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Study finds that eDNA can help detect rainbow smelt
Searun rainbow smelt—a culturally and ecologically valuable fish for New England anglers, consumers and marine ecosystems—is on the decline. Determining the extent of that decline, however, is difficult in Maine. Searun smelt can be easy to miss because they only enter coastal spawning streams from deeper waters during a few cold, wet nights each spring, and they depart the streams by early morning.
In a new study, a University of Maine-led research team found that collecting and analyzing environmental DNA (eDNA), fragments of DNA that organisms shed in their environment, can help detect the presence of rainbow smelt with greater accuracy, efficiency, safety and cost-effectiveness. It may also benefit surveys for other rare, transient fish species.
In response to the study findings, the Maine Department of Marine Resources will begin using eDNA and the UMaine-led team's tactics for employing it to survey several searun smelt streams in spring 2023.
Traditional monitoring methods for rainbow smelt involve either capturing them in streams with fyke nets or visually searching for spawning fish or eggs deposited on rocks. It can be challenging field work that grows harder as the fish become rarer. To obtain eDNA from rivers and streams, scientists can collect water samples in the daytime without specialized field gear or direct contact with the fish.
Researchers then use a process called quantitative polymerase chain reaction (qPCR), the same tool used to detect COVID-19, to isolate the DNA they want to study. Through laboratory analysis of water samples, qPCR detection of eDNA can reveal where a species has been, when they were at that location and how many were there at that time.
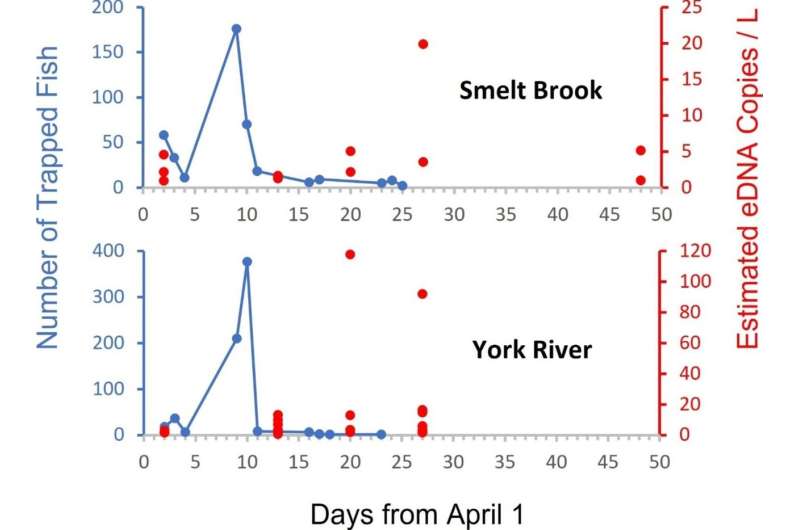
At the same time, eDNA can only be obtained in a limited amount of time after organisms leave a river or stream because the flowing water eventually dilutes and flushes it out of the system. Therefore, identifying optimal eDNA collection strategies for surveying rainbow smelt became part of the UMaine-led research team's investigation.
By surveying two sites in the York River system, the group found that eDNA samples were detectable eight–13 days after a single spawning event, a much longer window of time than that allotted for visual or net surveys. The team then surveyed at four sites around Casco Bay multiple times each in a month, after which they determined that employing three sampling events per location, three samples per event and six qPCR replicates per sample resulted in a greater than 90% combined detection capability.
"DMR is excited to apply eDNA techniques to help us take the pulse of smelt populations in Maine streams this coming spring," says Maine DMR scientist Danielle Frechette. "The results will complement the visual observations of smelt spawning being collected by community scientists along our coast and can help us identify locations for smelt restoration projects and better understand how climate change may be affecting smelt."
The study was led by Vaughn Holmes, who conducted the work when he was a master's student in ecology and environmental sciences, in collaboration with his graduate adviser Michael Kinnison, director of the Maine Center for Genetics in the Environment; Geneva York, manager of the UMaine Environmental DNA CORE Laboratory; and Jacob Aman, stewardship director of the Wells National Estuarine Research Reserve. The team's findings were published in BMC Ecology and Evolution.
"With expanded use of the methodology on the horizon, I'm eager to see eDNA become a mainstay in simplifying anadromous fish surveying efforts across Maine," Holmes says.
The study is part of the five-year Maine-eDNA program launched in 2019.
"eDNA has really taken off as a tool for natural resources monitoring and management," says Kinnison, co-principal investigator for Maine-eDNA. "It is not only a powerful survey tool, it is very accessible. Almost anyone can be trained to collect a water sample, and I am especially excited for how eDNA can make it possible for more people, from all walks of life, to participate in monitoring the species and habitats they care about."
More information: Vaughn Holmes et al, Environmental DNA detects Spawning Habitat of an ephemeral migrant fish (Anadromous Rainbow Smelt: Osmerus mordax), BMC Ecology and Evolution (2022). DOI: 10.1186/s12862-022-02073-y
Journal information: BMC Ecology and Evolution
Provided by University of Maine




















