This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Deciphering disease progression and cell processes with TIGER, in vivo and non-invasively
Could patients in the future simply ingest a diagnostic probiotic based on programmed ribonucleic acids to analyze their intestinal health from individual cells? Researchers at the Helmholtz Institute for RNA-based Infection Research (HIRI) and the Julius-Maximilians-Universität (JMU) in Würzburg have developed a new technology they call TIGER. It allows complex processes in individual cells to be deciphered in vivo by recording past RNA transcripts. The findings were published in the journal Nature Biotechnology on January 5, 2023.
Bacterial and viral infections can cause severe acute symptoms, but they can also have devastating long-term consequences, such as triggering cancer. Consequently, scientists are looking for new approaches and technologies to better understand the course of diseases and predict the development of cells and tissues. They use increasingly precise methods to analyze the underlying processes in individual cells. One goal is to detect changes in gene activity, which in turn may indicate a pathological event.
Ribonucleic acids (RNAs) make an important contribution to this understanding. They can provide evidence of genetic activity as only active genes produce RNA copies (transcripts) in a process called transcription. However, RNA molecules expressed in transcription represent a current state only. Linking past cell events—for example, a bacterial infection—to present conditions and deducing future outcomes proves challenging.
"The identity and behavior of a cell not only depends on its current intracellular make-up and extracellular environment but also on its past states. We were looking for an efficient procedure at the single-cell level to peer into the past and connect it to the present," explains Prof Chase Beisel, head of the RNA Synthetic Biology Department at the Helmholtz Institute for RNA-based Infection Research (HIRI) in Würzburg and the lead author of the study.
Peering into the cellular past
In their publication, the authors present a new technological approach that could significantly advance medical diagnostics in the future. Called TIGER, their method is a way to record the presence of specific RNAs in individual living cells.
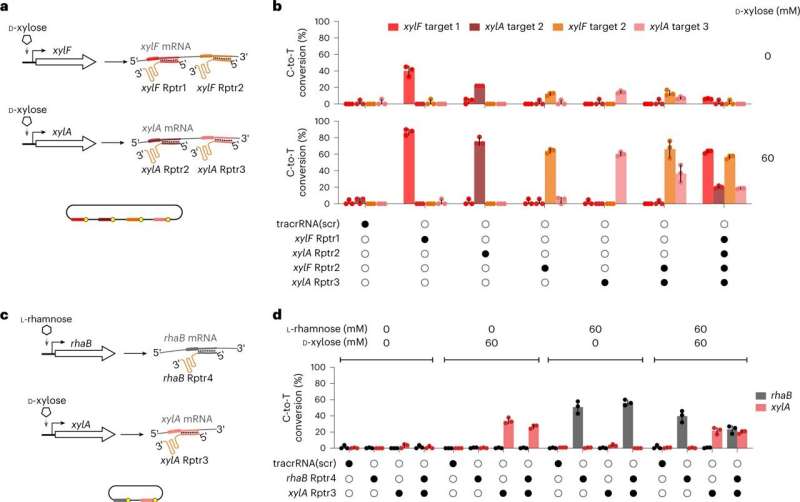
"Through RNA recording, TIGER connects current cellular states with past transcriptional states," says first author Chunlei Jiao. TIGER can quantify relative gene expression, detect differences between individual nucleotides, record multiple transcripts simultaneously and read out single-cell phenomena.
According to Jiao, there are striking advantages to the method: "Previous research has only been able to approximate past cell states using huge amounts of data and computational prediction tools to measure asynchronous cells over time." The scientists involved in the study were able to record the transfer of antibiotic resistance between Escherichia coli cells as well as the invasion of host cells by Salmonella.
In the future, TIGER could be used to look into the transcription history of individual cells in a living organism and link it to the current status quo in order to decipher complex cellular reactions—in vivo and non-invasively. For example, one could imagine ingesting a TIGER probiotic to have the state of the digestive tract recorded and analyzed later on, the authors conclude.
TIGER (an acronym of "Transcribed RNAs Inferred by Genetically Encoded Records") uses reprogrammed tracrRNAs (Rptrs) to record selected cellular transcripts as stored DNA edits in individual living bacterial cells. Rptrs are designed to base pair with recognized transcripts and convert them into guide RNAs.
The guide RNAs then instruct a Cas9 base editor to target an introduced DNA target. The extent of base editing can then be read by sequencing. The technology takes advantage of findings that led to the development of LEOPARD, an in vitro diagnostic platform, in a previous study.
More information: Chunlei Jiao et al, RNA recording in single bacterial cells using reprogrammed tracrRNAs, Nature Biotechnology (2023). DOI: 10.1038/s41587-022-01604-8
Journal information: Nature Biotechnology
Provided by Helmholtz Association of German Research Centres