Effect of chain length on adsorption. (A) AFM-based single-molecule imaging for DNA12kbp, DNA24kbp, and DNA48kbp on mica substrates observed in 10Ni. Images were acquired after incubating the mica substrates with 3.1 pM dsDNA solution in 5Ni for 2 min. (B) Relationship between Lc and Lex. Dashed line indicates a match between Lc and Lex. (C) Plots show the relationship between Nbp and Rg,2D. The blue line corresponds to the best fit of the experimental data assuming Lp,2D of 48 ± 7 nm. For comparison, dashed line superimposes Rg in 5Ni. (D) [Ni2+] dependence of Rg,2D for DNA48kbp. For comparison, the dotted line superimposes Rg in 5Ni. (E) AFM images for DNA12kbp and DNA48kbp acquired under the same condition as those for (A), except the incubation time was increased to 30 min. Credit: Science Advances (2022). DOI: 10.1126/sciadv.abn6349
Macromolecules in diverse phases can adsorb onto natural systems, composite materials, and thin-film devices. In a new report now published in Science Advances, Yuma Morimitsu and a research team in applied chemistry and polymer interfaces and molecular adhesion used atomic force microscopy (AFM) to visualize well-defined double-stranded DNAs and conducted molecular dynamics simulations to identify the adsorption mechanisms of polymer chains to solid surfaces.
The study outcomes can contribute to the development of light and durable polymer composites and films across a variety of industrial, biomedical and environmental applications.
The adsorption behavior of string-like molecules
Homeostasis of life involves natural systems of biomolecules such as proteins or nucleic acids that can adsorb onto a solid interface. This has been a topic of interest in polymer engineering fields for the past several decades. For instance, the polymer layer adjacent to a solid surface has a distinct structure from that of the bulk, where their aggregation states and thermal molecular motion can completely differ, compared to those in the bulk, and are therefore critical for the performance of polymer composites and thin-film devices.
Dip and spin-coating methods are classic examples of the physisorption of polymer chains from a solution system to adsorb macromolecules on to a surface. In this work, Morimitsu and colleagues directly viewed three double-stranded DNAs with different lengths via atomic force microscopy (AFM) to provide information on the adsorption behavior of string-like molecules from a solution to a solid surface.
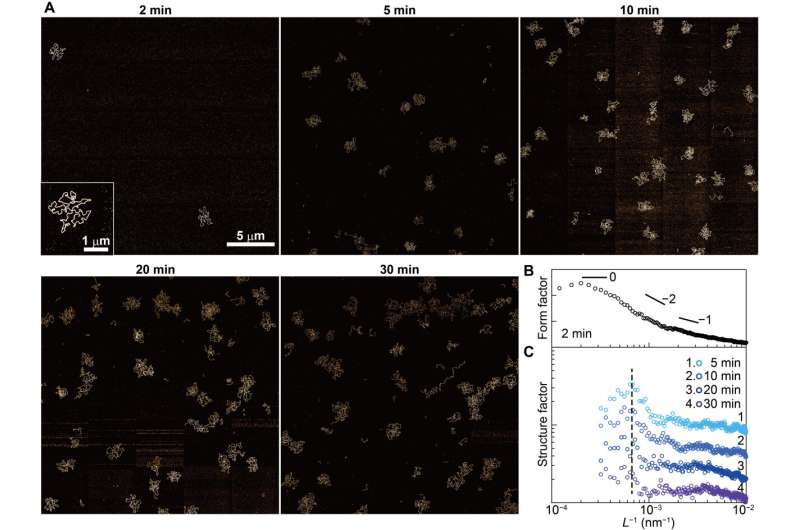
Cooperative adsorption of dsDNA. (A) AFM images for DNA48kbp adsorbed on mica acquired in 10Ni after incubating with a 3.1 pM DNA48kbp solution in 5Ni for 2, 5, 10, 20, and 30 min at 304 K. Inset is an enlarged image. (B) Average FFT power spectrum obtained from six AFM images acquired after incubating for 2 min. This spectrum was used as the form factor. (C) Spectra of the structure factor obtained by dividing the FFT spectra for 5 to 30 min by the form factor. Peak around 6.7 × 10−4 nm−1 corresponds to the distance between the adsorbed dsDNA chains on mica. The calculated value is 1.5 μm. Credit: Science Advances (2022). DOI: 10.1126/sciadv.abn6349
The initial adsorption of double-strand DNA chains
During the study, the research team directly observed single molecules to understand the extension of double-stranded DNA (dsDNA) on a solid. To accomplish this, they examined the chain dimension of double-stranded DNA chains from a solution onto a solid.
In this instance, the DNA chains were sandwiched between a hard wall and liquid molecules, where a side of the chain was in contact with a solid substrate, while the other was in contact with a solution. The outcomes highlighted the potential of DNA chains to adsorb onto mica and other substrates. Next, the team observed the mechanisms of cooperative adsorption, where the partially adsorbed chains were in an intermediate state, while some segments sought adsorption sites on the solid.
Formation of the interfacial layer
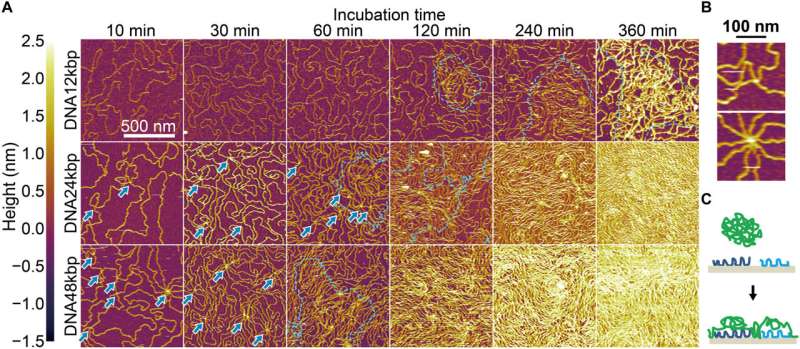
The researchers examined the formation of an interfacial layer on a solid thereafter, and obtained atomic force microscopy (AFM) images for specimens after incubating double-stranded DNA solution in buffer containing nickel for different periods of time. As they increased the time of incubation, the amount of adsorbed DNA chains increased. By 10 to 60 minutes, the mica substrate was covered with DNA, while many strands overlapped on account of cooperative adsorption. The researchers noted increased chain density at approximately 120 to 140 minutes.
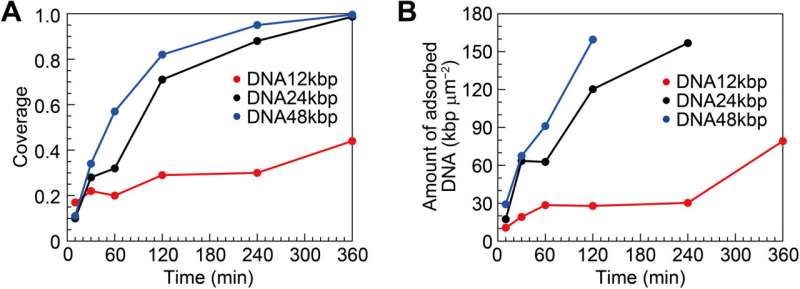
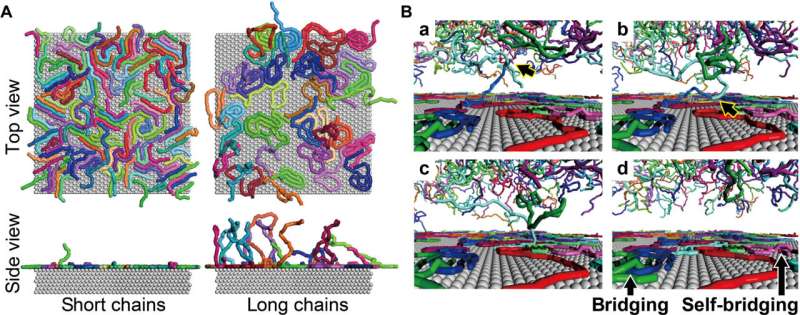
The team quantified the amount of DNA adsorbed on mica to show the surface coverage and adsorption analytics of DNA as a function of the time of incubation. They estimated the amount of DNA adsorbed on the mica surface and that floating in the buffer solution. During cooperative adhesion, the process of the adsorption of string-like molecules to solid surfaces became faster. They used coarse-grained molecular dynamics simulations to verify the hypothesis of DNA chain adsorption.
The team later studied the influence of the aggregation states of interfacial layers on the mechanical properties of composite materials. They accomplished this via a model of bulk films made of DNA and hydrated choline dihydrogen phosphate, which was rubber-like at room temperature, and subsequently measured their tensile properties.
-
Formation of interfacial layers. (A) AFM images for three kinds of dsDNA chains acquired after incubating mica substrates with 62.5 pM dsDNA solution in 2.5Ni for 10 to 360 min and subsequently washing with water. (Top) DNA12kbp, (middle) DNA24kbp, and (bottom) DNA48kbp. (B) Enlarged images for (top) DNA24kbp and (bottom) DNA48kbp to clarify the overlapping points. Images were acquired after incubating for 10 min. (C) Schematic illustration of the proposed bridging adsorption. Credit: Science Advances (2022). DOI: 10.1126/sciadv.abn6349
-
Surface coverage and amount of DNA for interfacial layers. (A) Surface coverage and (B) amount of DNA as functions of incubation time estimated from the AFM images. Credit: Science Advances (2022). DOI: 10.1126/sciadv.abn6349
-
Snapshots of chain adsorption by CGMD simulations. (A) Top and side views of adsorbed chains for the short- and long-chain systems with a surface coverage of 65%. (B) Characteristic scenes of cooperative adsorption observed in the long-chain system. Bridging adsorption also occurs. Credit: Science Advances (2022). DOI: 10.1126/sciadv.abn6349
Outlook
The team observed the dynamics of double-stranded DNA in solution, while noting their free existence. Thereafter, they noted how a segment of the DNA chain interacted with a solid surface to create a partially adsorbed segment. Yuma Morimitsu and colleagues used AFM (atomic force microscopy)-based direct visualization to reveal the dimension of polymer chains adsorbed on a surface and compared the dynamics to that in a solution state.
Based on the simulations, they revealed that unlike small molecules that adhered to Langmuir's hypothesis; in which a given surface has a number of sites for a species to stick via chemisorption or physisorption, the DNA polymers adsorbed on the surface in an isolated and cooperative manner. During the work, the team observed a narrow dispersion of double-stranded DNAs to provide insight to the macromolecular picture.
The outcomes can lead to the design of interfacial layers and the formation of polymer-based composite materials for applications across industrial, environmental and biomedical fields.
More information: Yuma Morimitsu et al, Direct visualization of cooperative adsorption of a string-like molecule onto a solid, Science Advances (2022). DOI: 10.1126/sciadv.abn6349
Kumaran S. Ramamurthi et al, Geometric Cue for Protein Localization in a Bacterium, Science (2009). DOI: 10.1126/science.1169218
Journal information: Science Advances , Science
© 2022 Science X Network
![Effect of chain length on adsorption. (A) AFM-based single-molecule imaging for DNA12kbp, DNA24kbp, and DNA48kbp on mica substrates observed in 10Ni. Images were acquired after incubating the mica substrates with 3.1 pM dsDNA solution in 5Ni for 2 min. (B) Relationship between Lc and Lex. Dashed line indicates a match between Lc and Lex. (C) Plots show the relationship between Nbp and Rg,2D. The blue line corresponds to the best fit of the experimental data assuming Lp,2D of 48 ± 7 nm. For comparison, dashed line superimposes Rg in 5Ni. (D) [Ni2+] dependence of Rg,2D for DNA48kbp. For comparison, the dotted line superimposes Rg in 5Ni. (E) AFM images for DNA12kbp and DNA48kbp acquired under the same condition as those for (A), except the incubation time was increased to 30 min. Credit: Science Advances (2022). DOI: 10.1126/sciadv.abn6349 Directly visualizing the cooperative adsorption of a string-like molecule onto a solid with double-stranded DNA](https://scx1.b-cdn.net/csz/news/800a/2022/directly-visualizing-t.jpg)