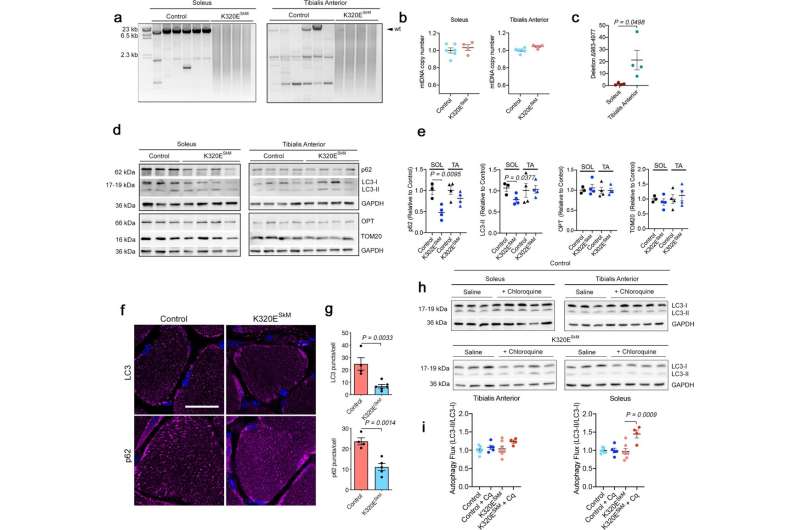
In vivo expression of Twinkle-K320E induces differential accumulation of mtDNA alterations. a Long range PCR analysis and b quantitation of mtDNA copy number in M. soleus and M. Tibialis anterior from 24 months old control and Twinkle-K320ESkM mice (control: n = 6; K320E: n = 4). c qPCR quantitation of deletion mtDNA-Δ983-4977 in M. soleus and M. Tibialis anterior from 2 years old mice (n = 4). d, e Western blot analysis and quantification of the indicated proteins in muscle extracts from these mice. (Soleus, control: n = 3; Twinkle-K320E: n = 4. M. TA, control: n = 4; Twinkle-K320E: n = 4). f, g In situ immunofluorescence and image quantification showing autophagic markers LC3 and p62 in cryosections of M. soleus. 5 random pictures with 4 fibers per plane were analyzed per animal to obtain averaged values (control: n = 4; K320E: n = 5). Scale bar, 20 µm. h, i Autophagic flux analysis (LC3-II/LC3-I ratio) in muscle extracts from mice treated with saline or 50 mg/kg chloroquine (Cq) 4 hours before euthanisation. (control, n = 7; control+Cq, n = 5; K320E, n = 7; K320E + Cq, n = 4). P values calculated using unpaired two-tailed Student’s t test (c, e), or One-way ANOVA with Tukey correction for multiple comparison (h). Data are presented as Mean ± SEM. Credit: Nature Communications (2022). DOI: 10.1038/s41467-022-34205-9
Scientists at the University of Cologne have discovered how cells can eliminate mutated mitochondrial DNA (mtDNA). Due to their evolutionary descent from bacteria, mitochondria still have genetic material packaged in chromosome-like structures (nucleoids). They convert the chemical energy in our food into a biologically usable form.
A team of researchers from the University of Cologne's Physiology Center at the Faculty of Medicine, the Center for Molecular Medicine Cologne (CMMC) and the CECAD Cluster of Excellence for Aging Research has now shown that mutations of the mtDNA lead to a local rearrangement of proteins in the mitochondrial membrane.
The mutated mtDNA is targeted, eliminated, and subjected to autophagy, the cellular waste disposal. The results have appeared in Nature Communications under the title "Mitochondrial membrane proteins and VPS35 orchestrate selective removal of mtDNA."
In many tissues, mutations in mtDNA accumulate as a result of normal aging. These kinds of mutations are an important cause of many aging-associated diseases. There are thousands of copies mtDNA in every cell, so mitochondrial function is only impaired when the percentage of mutated mtDNA molecules exceeds a certain threshold value.
It has long been established that mitochondrial damage, including acute mtDNA damage, triggers the process of mitophagy. In this process, dysfunctional mitochondrial parts are selectively degraded and recycled.
Dr. David Pla-Martin, the lead author of the current study, explained the details: "What is new in our study is that this mechanism does not affect the cells' endowment with mitochondria, but only clears out the damaged mtDNA. By labeling neighboring proteins—so-called proximity labeling—we showed that mtDNA damage leads to the recruitment of endosomes in close proximity to nucleoids."
Their removal is coordinated by the interaction of the nucleoid protein Twinkle and the mitochondrial membrane proteins SAMM50 and ATAD3. ATAD3 controls their distribution, SAMM50 induces the release and transfer of the nucleoid to the so-called endosomes.
"This additionally prevents the activation of an immune response. The protein VPS35, the main component of the retromer, mediates the maturation of early endosomes into late autophagy vesicles, where degradation and recycling ultimately take place," said Pla-Martin.
Using a mouse model in which mtDNA mutations lead to impaired muscle regeneration, the scientists also showed that selectively mutated mtDNA can be removed by stimulating the activity of the autophagy mechanism with rapamycin. The total number of mtDNA copies thus remains constant, preserving mitochondrial function.
Group leader Professor Dr. Rudolf Wiesner added, "Mutations in the genes that code for these proteins lead to severe neurological diseases, in VPS35 for example Parkinson's disease. We now want to use these proteins as new molecular targets to open up entirely new treatment options for these kinds of aging-associated diseases."
Although the road to therapeutic application could still be long, the research team sees a promising approach here.
More information: Ayesha Sen et al, Mitochondrial membrane proteins and VPS35 orchestrate selective removal of mtDNA, Nature Communications (2022). DOI: 10.1038/s41467-022-34205-9
Journal information: Nature Communications
Provided by University of Cologne