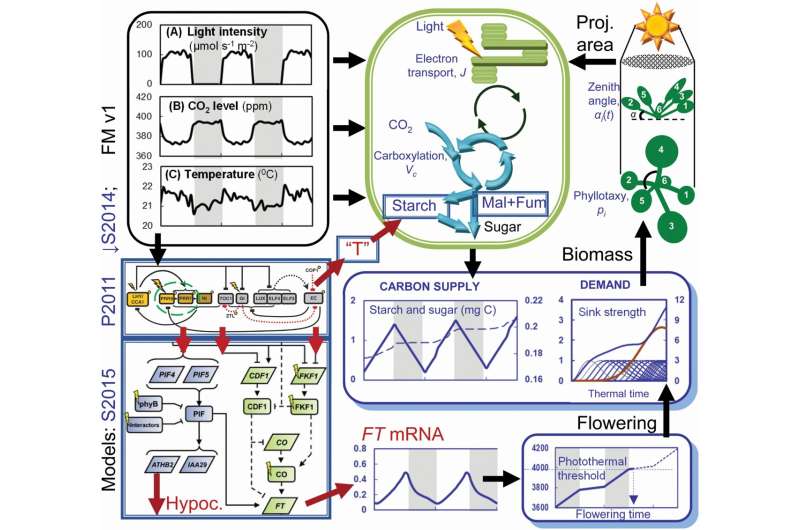
Operation of the Arabidopsis Framework Model version 2 (FMv2). Model components that are updated in FMv2 are bounded in double rectangles, with abbreviated model names (left). Key outputs from the new models are shown as arrows leaving the bounding rectangles. The P2011 clock model shows only clock genes for clarity. Morning or day genes, yellow symbols; evening genes and the evening complex (EC), gray symbols. The PRR9 and PRR7 genes that are inactivated in the prr7prr9 double mutant are marked with a dashed oval. Components of P2011 drive the S2014 starch degradation model via component ‘T’ (center), and the S2015 external coincidence model (lower left). S2015 controls photoperiod-dependent hypocotyl elongation (Hypoc.) by rhythmically gating the PhyB/PIF/ATHB2 pathway, and the florigen FT by the CDF1/FKF1/CO pathway. The S2015 cartoon distinguishes RNA components (paralellograms) from proteins (rounded rectangles). Light inputs are shown as flashes. Among the model components retained from the FMv1, the carbon dynamic model (bounded in green) includes updated starch and malate and fumarate (Mal + Fum) carbon stores, and provides Sugar as the Carbon Supply for growth. This is allocated according to the Demand of leaf (blue) and root (red) sinks in the functional–structural plant model, which uses the leaf Biomass to calculate the rosette’s projected area for photosynthesis (Proj. area). When the Photothermal model (lower right) reaches the threshold for Flowering, the simulation ends. Credit: in silico Plants (2022). DOI: 10.1093/insilicoplants/diac010
Scientists have made a significant step towards building the world's first digital plant by developing a sophisticated computational model which has also solved one of the most enduring plant science mysteries—the role of the biological clock.
Anyone who has suffered from jet lag can identify with the devastating effects of a disrupted biological clock. Now a new study has revealed that plants also suffer when their clocks are out of sync.
By creating a permanently jet lagged plant—the equivalent of flying from New York to the UK every day—researchers discovered that disrupting a plant's biological clock impacts their growth. The team also created a computer model of the "jet-lagged" plant that was able to accurately predict the effect on growth—and reveal which molecular pathways are impacted by faulty clock genes.
The advance represents a significant step forward in creating a complex, multicellular digital organism—a feat which has rarely been achieved outside of single-celled microbes.
The approach, which has been discussed for over a decade, should soon extend to other clock-regulated pathways, and lead to fresh insights into wider plant biology that could help improve crop yields and resilience to better cope with climate change.
All plants have a biological clock, a molecular time-keeping system that detect shifts in the environment and prepares the plant for changes from dusk to dawn and season to season. Although every plant cell seems to have its own clock, which controls around 30% of its genes, little was known about their role in plant growth.
To tackle this a study, by researchers at the University of Edinburgh, investigated the effects of mutations in the clock genes of Arabidopsis thaliana, a widely studied plant species. The clock-mutant plants allowed the team to investigate whether clock genes were involved in the plant's nightly release of sugar stored in starch, which fuels their growth
Plants need to carefully manage the energy they capture through photosynthesis during daylight hours. Releasing sugar from starch stores too quickly or slowly overnight can stall their growth. Scientists studied the growth of Arabidopsis plants with mutations in their clock genes that caused the clock to run too slowly—as if the day was 29 hours rather than 24 hours long.
In these mutants, night-time release of sugars from starch was slower than in normal plants and growth was reduced.
They also created a computational model of these clock mutants, known as a Framework Model, which combined mathematical models of clock gene activity with metabolic and physiological models. The results revealed that the Framework Model accurately simulated the effects on plant growth—correctly predicting that the slow release of sugars from starch during the night in the clock mutants was responsible for slowing their growth
The findings contrast with earlier studies of other clock-mutants, which indicated that disrupted biological clocks interrupt plant growth by affecting key processes in photosynthesis. As well as revealing the role of the plant's 24-hour clock, the framework model was able to link the genes, through measurable molecular pathways, to its impact on the whole plant—a classical challenge in genetics.
This achievement is the equivalent to understanding a human health syndrome caused by a genetic change that subtly influences multiple, physiological pathways.
The team's next step is to use the Arabidopsis framework model to predict how the plant's genome sequence controls these physical characteristics and traits, known as its phenotype. If successful, the approach could be applied more widely and lead to the sought after "grand unified" understanding of biology—revealing the interplay between genomes and the living systems they create.
Using this approach, which aims to predict how living systems work, similar models could be developed to help make sense of the vast data sets generated by advances in genome sequencing. This type of advance could also unravel the complexity of molecular results to decipher which are the most important and have the greatest impact on health and disease in living organisms.
Professor Andrew Millar of the University of Edinburgh's School of Biological Sciences, says that "the success of the Framework model shows that we can understand subtle effects at the whole-plant level, in this case just from changing the timing of gene expression. By 'understand' we mean 'explain and predict'. Not all details of this model will transfer to crop species, but it extends the 'proofs of principle' for informing crop improvement at the molecular level."
The study was published in in silico Plants.
More information: Andrew Millar et al, The Arabidopsis Framework Model version 2 predicts the organism-level effects of circadian clock gene mis-regulation, in silico Plants (2022). DOI: 10.1093/insilicoplants/diac010
Provided by University of Edinburgh