How deep learning empowers cell image analysis
The cell is the basic structural and functional unit of life, with varying sizes, shapes, and densities. There are many different physiological and pathological factors that influence these parameters. It is therefore extremely important for biomedical and pharmaceutical research to study the characteristics of cells.
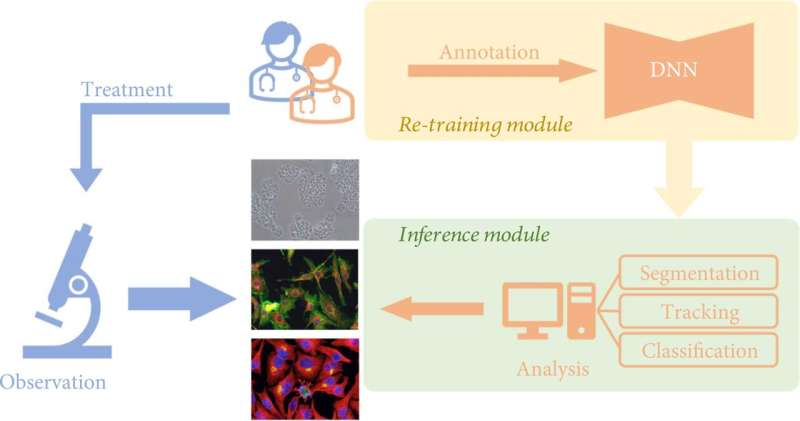
Traditionally, researchers observed cell samples directly through microscopes in order to study the morphological changes of cells. In recent years, with the development of computer science and artificial intelligence, deep learning can now be combined with cell analysis methods. This can replace researchers' direct observation under the microscope and manual interpretation of images, improving the efficiency and accuracy of research.
An increasing number of deep learning-based algorithms have been developed to empower cell image analysis, primarily for addressing three key tasks:
- Segmentation. To identify meaningful objects or features, the image is divided into several parts using deep learning. Cell segmentation is the basic premise of identifying, counting, tracking, and morphological analysis of cell images;
- Tracking. That is, after segmentation of the cell images, the cell behavior of the entire spectrum is monitored. Living cells contain a lot of information about the living organism, and the dynamic characteristics of cells, especially morphological changes, can reflect the health status of the organism in the pathological and physiological processes, such as immune response, wound healing, cancer cell spreading and metastasis, etc.
- Classification. The classification of cell morphological features based on extracted parameters often serves as a downstream analysis task for phenotypic screening and cell profiling.
For the above three crucial tasks, a review article published in the journal Intelligent Computing discusses in depth the progress of deep learning techniques.
"In contrast to traditional computer vision techniques, a deep neural network (DNN) can automatically produce more effective representations than handcrafted representations by learning from a large-scale dataset. In cell images, deep learning-based methods also show promising results in cell segmentation and tracking," the authors said. "Such successful applications demonstrate the ability of DNNs to extract high-level features and shed light on the potential capability of using deep learning to reveal more sophisticated life laws behind cellular phenotypes."
In addition, the authors also discuss the challenges and opportunities of deep learning methods in cell image processing. Authors said, "Deep learning has demonstrated an incredible ability to perform cell image analysis. However, there remains a significant performance gap between deep-learning algorithms in academic research and practical applications." There are currently challenges and opportunities in three aspects, namely data quantity, data quality, and data confidence:
- Deep Learning with Small But Expensive Dataset. Constructing a large-scale cell image dataset is a strenuous task. This is because cell images require knowledgeable biological experts to assign labels image by image. The scale of cell image datasets is often limited by the difficulty of annotation.
- Deep Learning with Noisy and Imbalanced Labels. The quality of the annotations of cell image datasets is highly dependent on the professional skills of humans, resulting in label noise and label imbalance. Label noise is introduced by assigning incorrect or incomplete labels to training images. Label imbalance is caused by the preference for annotation, where the numbers of labeled images for different classes are quite unbalanced.
- Uncertainty-Aware Cell Image Analysis. Uncertainty-aware learning is crucial for deep-learning applications in biological scenarios. It is impossible for a plain neural network to detect new phenotypes without a mechanism to reflect the confidence of classification results.
Using deep learning, scientists are exploring new technologies to improve cell image analysis. More effective solutions will be proposed in the future, and deep learning and biomedical research will be more closely integrated.
More information: Junde Xu et al, Deep Learning in Cell Image Analysis, Intelligent Computing (2022). DOI: 10.34133/2022/9861263
Provided by Intelligent Computing