Widespread variation of inherited retroviruses among Darwin's finches
Vertebrate genomes are repositories for retrovirus code that was deposited into germ line as inherited endogenous retroviruses during evolution. Researchers from Uppsala University and Princeton University now provide new findings about retroviral establishment and distribution among Darwin's finches. The findings are being published in Nature Communications.
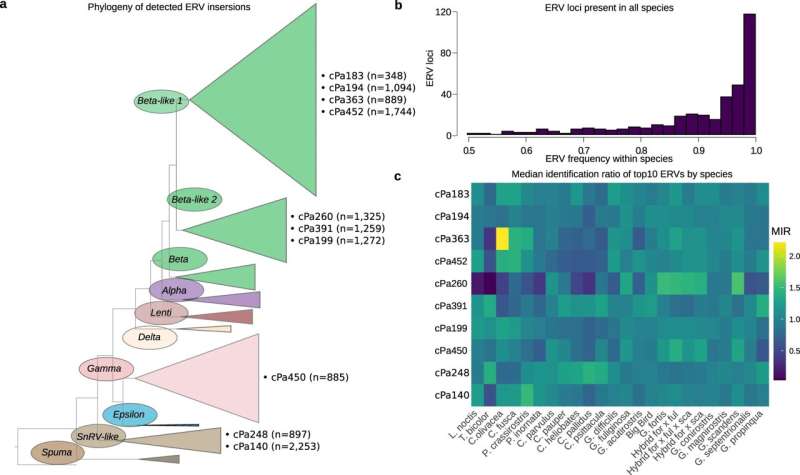
The researchers examined whole genomes sampled from the entire Darwin's finch radiation and found shared endogenous retroviruses (ERV) among all individual birds suggesting that most retrovirus-host interactions pre-date host speciation.
They also found considerable ERV variation across different populations of Darwin's finches suggesting more recent retrovirus colonization of germ line. Natural selection can then lead to enrichment or loss of ERVs. Occurrence is also affected by crosses between finch species, which results in gene flow including the ERVs.
"The unexpected ERV variation suggests recent retrovirus infection and historical changes in gene flow and selection," says Jason Hill, Uppsala University, shared first author of the study.
By mapping ERV variation across all species of Darwin's finches and comparing with related finch species, the researchers highlight geographical and historical patterns of retrovirus-host occurrence.
"The ERV distribution along and between chromosomes, and across the finch species, suggests connection between ERVs and the rapid speciation," says Mette Lillie, Uppsala University, shared first author of the study.
As a well examined species group which has become synonymous with evolution studies, Darwin's finches represent a natural model for evaluating the extent and timing of retroviral activity in hosts undergoing speciation and colonization of new environments.
"Darwin's finches provide a wonderful resource for connecting ERV variation across host populations with host genes and phenotypes, for identifying historic virus-host interactions and potential contributions to host biology," says Patric Jern, Uppsala University, who headed the study.
More information: Jason Hill et al, Spatiotemporal variations in retrovirus-host interactions among Darwin's finches, Nature Communications (2022). DOI: 10.1038/s41467-022-33723-w
Journal information: Nature Communications
Provided by Uppsala University