Developing self-complementary macrocycles with ingenious molecules
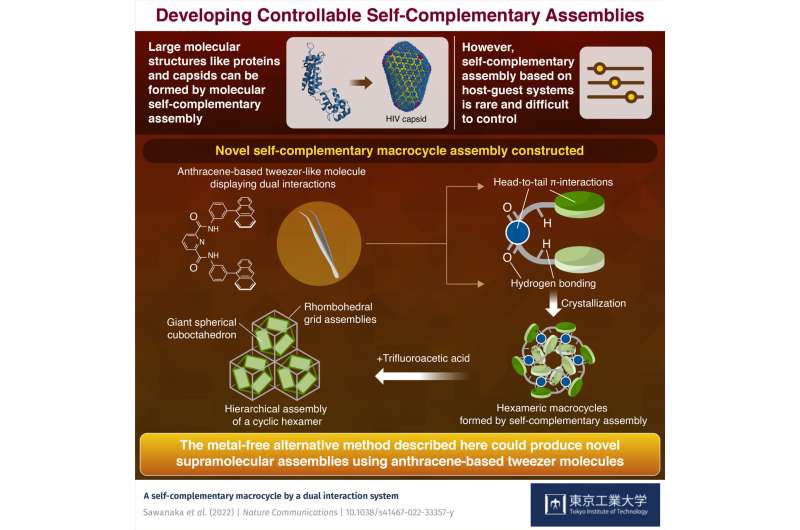
Some biological molecules with efficient noncovalent bonding sites can use their bonding properties to create well-defined assemblies from a single class of molecules—i.e., they assemble with each other. These molecules, which are frequently seen in nature, are referred to as "self-complementary assemblies." For instance, the p24 protein hexamer, which is part of the capsid of the HIV (human immunodeficiency virus), is composed of six protein subunits that complementarily self-assemble using many hydrogen bonds.
This phenomenon allows well-designed molecules to form higher-ordered assemblies without the metal ions that are commonly used as "joints" between monomer molecules. Indeed, many self-complementary assemblies have been reported on the basis of intrinsic hydrogen bonds, π-interactions, and coordination bonds.
However, self-complementary assembly based on host-guest systems is rare and notoriously difficult to control. In order to further our understanding of self-complementary assembly with higher-ordered structures, many new strategies have come to light in recent years.
Now, a team of researchers from the School of Science at Tokyo Institute of Technology (Tokyo Tech) might have just cracked the code to developing these innovative systems. The team, led by Assistant Professor Masahiro Yamashina and Professor Shinji Toyota, has constructed a novel self-complementary macrocycle assembly using an anthracene-based tweezer-like molecule with a pyridinedicarboxamide (PDA) linker as the monomeric species. Their work is described in Nature Communications.
"The molecule we use has an interesting property: It can bond with itself in two ways and form self-complementary structures. Not only does it show head-to-tail π-π interactions between the electron-rich tweezer tail (the anthracene groups) and the electron-deficient head, but also presents hydrogen bonding through the amide (-NH) functional group. By incorporating these two interactions, a preferential direction of self-assembly is achieved, and this guides the formation of the macrocycle," explains Prof. Yamashina.
This type of dual interaction leads to much more control over the formation of synthetic macrocycles, and in this case, gives rise to a stable self-complementary hexameric structure upon crystallization. These hexamers can further assemble into even bigger self-complementary structures in the right conditions.
"When we added trifluoroacetic acid (TFA), we found that the cyclic hexamers further assemble into two predominant, stable supramolecular structures: rhombohedral grid assemblies and giant spherical cuboctahedra, a so-called hierarchical assembly," Prof. Toyota says. "The latter structure is particularly impressive as it is formed from 108 monomeric tweezer units."
Current methods to form supramolecular assemblies require metals that could harm the environment and ecosystems. The metal-free alternative method described here could produce novel supramolecular structures using a simple anthracene-based tweezer molecule. It opens the door to a new range of supramolecular assemblies with optical and electronic functions. This work adds a new important tool in the chemistry toolbox, one that is sure to play a big role in the metal-free supramolecular structures of the future.
More information: Yuta Sawanaka et al, A self-complementary macrocycle by a dual interaction system, Nature Communications (2022). DOI: 10.1038/s41467-022-33357-y
Journal information: Nature Communications
Provided by Tokyo Institute of Technology