Scientists unravel millet genotype-microbiota interaction insights to enhance crop adaptability and yield
Foxtail millet, Setaria italica, is one of the oldest and more resilient crops worldwide. Compared to rice and wheat, millet has excellent climate resilience and requires less fertilizer, pesticides, and irrigation than mainstream cereals. In addition, millet-based foods are nutritionally superior to other cereal crops. With these properties, this crop is poised to play an important role to strengthen food security to feed the world's growing population.
BGI in collaboration with the Chinese Academy of Sciences has uncovered a millet plant genotype-microbiota interaction network that contributes to phenotype plasticity. This study was published on October 7, 2022 in Nature Communications.
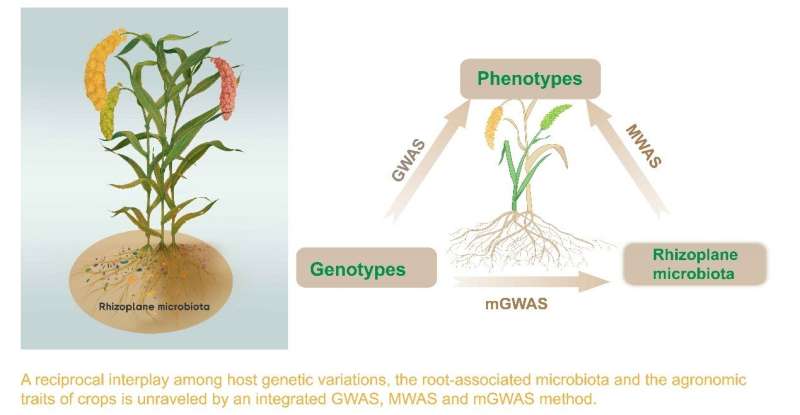
Previous studies revealed key loci for early and late flowering times and blast-resistance in foxtail millet, but the loci associated with plant growth or yield are still not known. This research revealed the association between millet genotype, root microbiome, and agronomic traits through association analysis, and proposed genotype-dependent microbial effects for the first time. This provides an in-depth analysis of the genetic and environmental factors that affect millet crop growth and yield.
The research was based on genetic variation data of 827 foxtail millet genomes of different varieties, rhizoplane microbiota (external root-surface) data, and 12 growth and yield phenotypic data of each millet. This study integrates genome-wide association studies (GWAS), microbiome-wide association studies (MWAS), and microbiome genome-wide association studies (mGWAS) methods to reveal associations between genotypes, agronomic phenotypes, and rhizoplane microbiota in foxtail millet.

The researchers identified 257 rhizoplane microbial biomarkers associated with six key agronomic traits. The rhizoplane microbiota composition is mainly driven by variations in plant genes related to immunity, metabolites, hormone signaling and nutrient uptake. Among these, the host immune gene FLS2 and transcription factor bHLH35 are widely associated with the microbial taxa of the rhizoplane. The microbial-mediated growth effects on foxtail millet are dependent on the host genotype, suggesting that precision microbiome management could be used to engineer high-yielding cultivars in agriculture systems.
These research results will help to enhance millet productivity and its adaptability to the environment. Yayu Wang, co-author and BGI senior researcher commented, "A 'personalized feeding strategy' with precision microbial biofertilizers will be a key to develop high-yielding cultivars in the future. This will involve integrating DNA-level precision breeding with microbial ecological fertilizers, water and fertilizer integration, full-field management, precision seeding and other modern planting technologies."
More information: Yayu Wang et al, GWAS, MWAS and mGWAS provide insights into precision agriculture based on genotype-dependent microbial effects in foxtail millet, Nature Communications (2022). DOI: 10.1038/s41467-022-33238-4
Journal information: Nature Communications
Provided by BGI Genomics





















