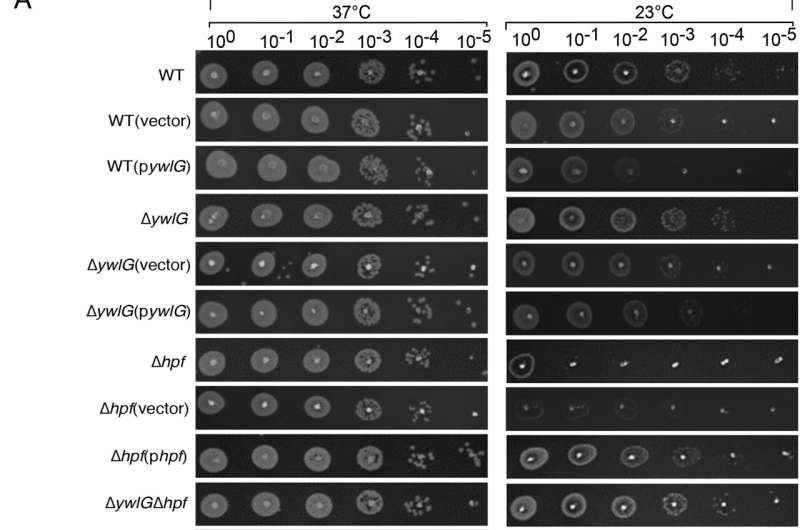
Inactivating ywlG suppresses the cold sensitivity of a Δhpf-null mutant in a medium-dependent manner. Complementation with plasmid-encoded ywlG and hpf confirmed the YwlG-mediated suppression and HPF-dependent cold response. Serial dilution spot assays were performed on (A) lysogeny broth (LB)-based or (B) tryptic soy broth (TSB)-based agar plates. Exponentially growing cells (OD600 = 0.6) in TSB were adjusted to OD600 = 0.2 after two washes in 1xPBS. After serial dilutions, 2 µL of each dilution was spotted on the agar plates. The results were recorded after 24 h and 48 h of incubation at 37 °C and 23 °C, respectively. The images are representative of three independent experiments. Credit: Proceedings of the National Academy of Sciences (2022). DOI: 10.1073/pnas.2207257119
Northwestern Medicine investigators have uncovered novel regulatory mechanisms that promote bacterial survival, according to findings published in the Proceedings of the National Academy of Sciences.
The study, led by M.-N. Frances Yap, Ph.D., associate professor of Microbiology-Immunology, also lays the groundwork for identifying new therapeutic targets to treat bacterial infections.
Ribosome hibernation is a mechanism carried out by bacteria and other organisms including eukaryotes to ensure long-term survival. Hibernating 100S ribosomes in bacteria consist of two 70S complexes joined together by a hibernation-promoting factor (HPF) protein.
This 100S complex serves two main purposes: to prevent the degradation of protein biosynthesis machinery (ribosomes) and to conserve energy in the cell by shutting down mRNA translation.
In the current study, Yap's team utilized mass spectrometry to study protein interactions in methicillin-resistant Staphylococcus aureus, a bacterium commonly known to cause staph infections. From these analyses, they found that HPF interacts with a previously unidentified binding partner located outside of the ribosome, a protein called YwlG.
Additionally, by solving the atomic structure of YwlG, they found this protein is involved in glutamate hydrogenase activity, which is an essential metabolic pathway required for bacteria survival.
"Under normal conditions, a fraction of YwlG protein is bound to the HPF. By binding together, YwlG cannot stimulate glutamate hydrogenase activity and HPF cannot bind to the ribosome, so it's 'mutual sequestration' of the two proteins to ensure the right amount of 100S complexes and cellular glutamate dehydrogenase is produced," Yap said.
The findings point to both HPF and YwlG as factors that influence bacterial colonization and infection severity. The end goal, according to Yap, is to identify a compound that can disrupt the formation of these complexes and, in turn, inhibit long-term survival of bacteria.
"One key advantage is that protein similar to HPF or YwlG does not exist in humans, so you could develop a HPF- or YwlG-targeting antibacterial that inhibits bacterial growth but does not harm the hosts," Yap said.
More information: David Ranava et al, Bidirectional sequestration between a bacterial hibernation factor and a glutamate metabolizing protein, Proceedings of the National Academy of Sciences (2022). DOI: 10.1073/pnas.2207257119
Journal information: Proceedings of the National Academy of Sciences
Provided by Northwestern University