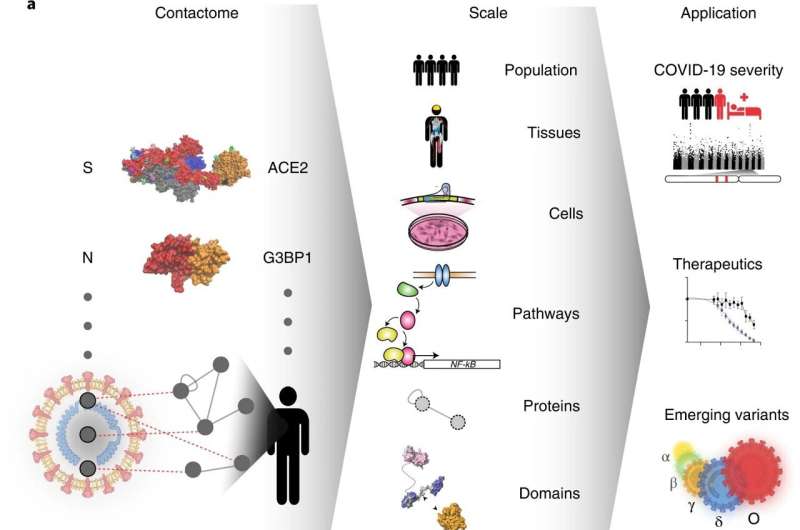
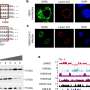
Generation and quality assessment of HuSCI. a, The contactome, the sum of physical contacts between viral and host macromolecules, mediates cellular perturbations that enable viral replication and cause disease manifestations. N, nucleocapsid protein; S, spike protein. b, Orthogonal validation; fraction of pairs that are yN2H-positive in HuSCI (top, n = 282 pair configurations representing 148 HuSCIs interaction pairs) and IntraSCI (bottom, n = 41 pair configurations for 25 IntraSCI interaction pairs), in the benchmark positive control sets hsPRS-v2 (n = 180 pair configurations for 60 interaction pairs) and vhLit-BM (n = 164 pair configurations for 40 interaction pairs), and negative control sets hsRRS-v2 (n = 234 pair configurations for 78 protein pairs) and vhRRS (n = 360 pair configurations for 178 protein pairs). Asterisks indicate significant differences from vhRRS benchmark (*P = 0.023; **P = 0.005; ****P = 1.57 × 10−5 hsPRS-v2, P = 1.02 × 10−7 HuSCI; NS, not significant; two-sided Fisher’s exact test; center, proportion of positives; error bars, standard error of proportion). c, Overlap of SARS-CoV-2 targets identified in HuSCI with previously identified target proteins of other viruses (left) and actual overlap (arrow) compared to n = 10,000 randomized control networks (right) (one-sided, empirical P < 0.0001). d, Host targets identified in HuSCI overlap with RNA-binding proteins (RBPs) bound to SARS-CoV-2 RNA upon infection (left) and actual overlap (arrow) compared to n = 10,000 randomized control networks (right) (one-sided, empirical P = 0.007). Credit: Nature Biotechnology (2022). DOI: 10.1038/s41587-022-01475-z
What exactly are the molecular interactions between the virus causing COVID-19 and its human host? How might our genetic differences cause different disease courses? And how do still-emerging virus variants differ in their host-virus interactions? To get to the bottom of these questions, an international team of researchers has generated a systematic map of molecular contacts between the SARS-CoV-2 virus and its human host.
The contact map, published in the journal Nature Biotechnology, reveals more than 200 direct protein-protein contacts, or protein interactions. The international consortium of scientists, led by Pascal Falter-Braun, Director at the Helmholtz Munich Institute of Network Biology (INET) and Professor at the Faculty of Biology, Ludwig-Maximilians-University (LMU) Munich, Germany, included teams in Canada, U.S., France, Spain and Belgium.
In contrast to previous large-scale studies on protein-protein associations, now the direct protein contacts between virus and host could be precisely identified. "To really understand the mechanistic connections between virus and host, we need to know how the parts fit together," says Frederick Roth, Professor at the University of Toronto's Donnelly Centre and Sinai Health (Toronto, Canada).
Taking a closer look at this newly-revealed set of direct protein interactions (or "contactome"), the team found chains of connections between viral proteins and infection-relevant human genes. For example, they were able to trace connections between certain SARS-CoV-2 proteins and human proteins encoded by the genes that have been linked to an increased likelihood of severe COVID-19 in other studies. They also found connections between the viral proteins and genes involved in, for example, metabolic disorders like obesity and diabetes.
"We already know that genetic differences in humans have an important role in the course and severity of a COVID-19 infection," says Pascal Falter-Braun, and he continues, "thanks to identification of the molecular contact points, it is now possible to examine the underlying mechanisms."
Initial insights included a demonstration that important inflammatory signaling pathways are directly activated by the virus. These contacts may help explain the exaggerated inflammatory reaction, that plays a major role in severe cases of COVID-19.
However, the protein-protein contacts not only point to impacts on the function of human cells and the human immune system, but also impact the function of SARS-CoV-2, including the rate at which the virus replicates.
According to Falter-Braun, the interaction of virus and human cells can be thought of as a visit of the virus to a restaurant: The guest—the virus—initially only has contact with the waiter, but subsequently the waiter goes to the kitchen, communicates the order to the chef, and the virus again gets an answer, in this example the meal, which in turn affects the virus. Depending on which proteins in the human cells—that is: waiter, chef, kitchen assistant, etc—encounter which proteins of the virus, the infection and immune reaction can turn out differently.
"Because of this mutual influence of protein-protein connections, our systematic contact map points to many potential drug targets," says Falter-Braun. The scientists have already been able to confirm, for example, that the human protein USP25 is recruited to help certain viral process and that its inhibition significantly reduces the multiplication of the virus.
"Many of the technologies and collaborations in this study were developed for other purposes, then quickly 'pivoted' to the COVID-19 pandemic, highlighting the value of fundamental research investments" says Dr. Dae-Kyum Kim, a lead author who began this work at Sinai Health (Toronto) and continued it as an Assistant Professor at the Roswell Park Comprehensive Cancer Center.
To this end, they first had to put in some effort and use the latest technology, because charting the contact map was at times like solving a huge puzzle for the international team of researchers: The scientists systematically examined and displayed the interactions of around 30 viral proteins, each with ~17,500 human proteins in so-called assays, interrogating 450,000 protein pairs.
By hand, one would never have managed this in such a short time. "We resorted to robotics when testing the individual plates each with multiple assays so that each protein type was automatically paired with another. And we had artificial intelligence methods do the initial evaluation of whether interactions occurred or not," Falter-Braun says.
Such a mammoth project required a team effort. "From molecular biology methods to computational analysis of networks and protein domains, to expertise in virology and innate immunity, we collaborated across disciplines," says Falter-Braun. "Our expertise in virus-host interactomics combined with RNA viruses biology, enabled assessing the virus' dependence on host direct partners" specifies Caroline Demeret from the Institut Pasteur.
The effort, researchers believe, was worth it: The contactome map will serve as a platform for the scientific community to study individual interactions in more detail and to understand their impact on molecular mechanisms and clinical progression, and thus uncover starting points for new therapeutic possibilities.
More information: Caroline Demeret, A proteome-scale map of the SARS-CoV-2–human contactome, Nature Biotechnology (2022). DOI: 10.1038/s41587-022-01475-z
Journal information: Nature Biotechnology
Provided by Helmholtz Association of German Research Centres