Graphical abstract. Credit: Molecular Cell (2022). DOI: 10.1016/j.molcel.2022.06.031
For four decades, Specificity Protein 1 (Sp1) has been cast solely as a transcription factor, a type of protein that binds DNA and turns genes on. But a Donnelly Center study has now revealed that Sp1 plays another unexpected role in gene regulation by influencing the stability of genes' RNA messages.
The finding is especially significant for cancer research, as it upends the established thinking about how Sp1 contributes to the disease and opens new avenues for treatment development.
"Sp1 has been known to be involved in cancer for a long time, but it was thought to be because of its ability to bind DNA and act as a transcription factor, because everybody knew about that," said Jack Greenblatt, senior author on the paper and a professor of molecular genetics in the Donnelly Center for Cellular and Biomolecular Research, at U of T's Temerty Faculty of Medicine.
"We think that's not the answer. We think it affects cancer thorough its ability to bind RNA and regulate transcript stability," said Greenblatt, who is also a University Professor.
The journal Molecular Cell published the findings.
The research sheds unexpected new light on one of the most studied human proteins. Sp1 was first identified as a DNA-binding transcriptional activator nearly 40 years ago and was the first human transcriptional regulator to be purified, said Greenblatt. "It is a standard topic in undergraduate textbooks," he added.
Sp1 controls activation of around 6,000 genes—roughly a third of the human genome—which are mostly required for cellular sustenance.
And now there's also the RNA-binding Sp1, which holds sway over another and mostly non-overlapping group of 2,000 genes by impacting abundance of their mRNA copies, according to the study.
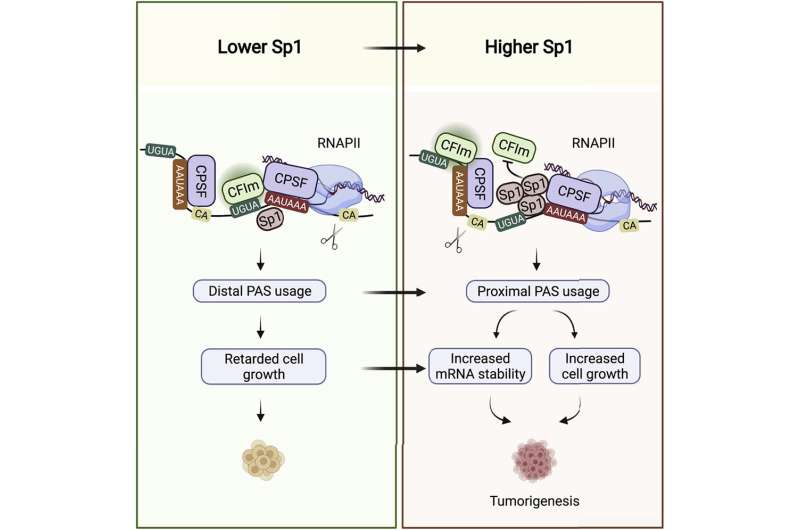
When a gene is switched on, its code gets copied into a messenger RNA, or a transcript, which serves as a template for building the encoded protein molecule. At its tail end, mRNA harbors the untranslated region, or UTR, which does not get translated into protein, but which has a regulatory role and is important for transcript stability. Research has shown that transcripts with long UTR tails are often less stable and get degraded more rapidly by cellular enzymes, while short-tailed UTRs are more protected from degradation.
"It is here, in the UTR, that Sp1 binds its target mRNAs," said Syed Nabeel-Shah, a Ph.D. candidate in the lab and the research's co-first author, along with former Greenblatt laboratory post-doctoral fellow, Jingwen Song. When Nabeel-Shah and Song depleted Sp1 from cells, they found that this led to an increase in certain transcripts' UTR length, indicating that Sp1 acts to yield shorter UTR tails. They further showed that UTR trimming is accomplished by the RNA cleavage machinery, which snips the RNA near where Sp1 is bound.
Control of UTR length has emerged as an important layer of gene regulation. This is especially true in cancer cells, which abound in short-tailed transcripts. The shorter the UTR tail, the more stable the mRNA for many transcripts, which means more protein templates—and ultimately more protein molecules that can be produced.
"Fast-growing cells need certain genes expressed at higher levels," said Greenblatt. "The effect of having shorter UTRs is often a higher concentration of mRNA, and consequently higher protein concentration."
It remained unclear, however, how balance is skewed towards shorter UTRs in cancer cells. But Greenblatt and Nabeel-Shah think that Sp1 holds part of the key.
It's long been known that Sp1 levels are elevated across many types of cancer, including breast cancer, which Nabeel-Shah and Song focused their analysis on. They obtained RNA sequencing data from the Cancer Genome Atlas on one thousand breast cancer patients. They found that Sp1 levels correlate with its target transcript tail length and abundance. The more Sp1 in a patient sample, the shorter the UTRs in the same sample and the higher the mRNA levels when those UTRs are bound by Sp1. "All this goes to suggest that Sp1 promotes cancer through its RNA-binding role," said Nabeel-Shah.
The finding is in contrast with the established view where Sp1 contributes to cancer by turbocharging the expression of its target genes as a DNA-binding protein. Nabeel-Shah and Song found no support in the data for this concept. There was no significant correlation between Sp1 levels in breast cancer cells and the amount of mRNA transcribed from the genes it targets on DNA.
Although they only looked at breast cancer, the researchers think that Sp1 plays a similar role in other types of cancer, given the large number of mRNAs it regulates.
The study opens up a new way of thinking not only about Sp1 and its role in cancer, but also about other similar proteins. Sp1 belongs to a family of C2H2 zinc finger transcription factors, which form the largest group of transcription factors in humans. The majority of its over 700 members remain poorly explored, however.
The goal of Nabeel-Shah's thesis is to determine if a lot of other C2H2 proteins also bind RNA. So far, he has tested 150 of these proteins and found that 145 bind both DNA and RNA, suggesting that many other members too might have dual roles in gene regulation.
Sp1 was only the first to be discovered, just like four decades ago.
More information: Jingwen Song et al, Regulation of alternative polyadenylation by the C2H2-zinc-finger protein Sp1, Molecular Cell (2022). DOI: 10.1016/j.molcel.2022.06.031
Journal information: Molecular Cell
Provided by University of Toronto