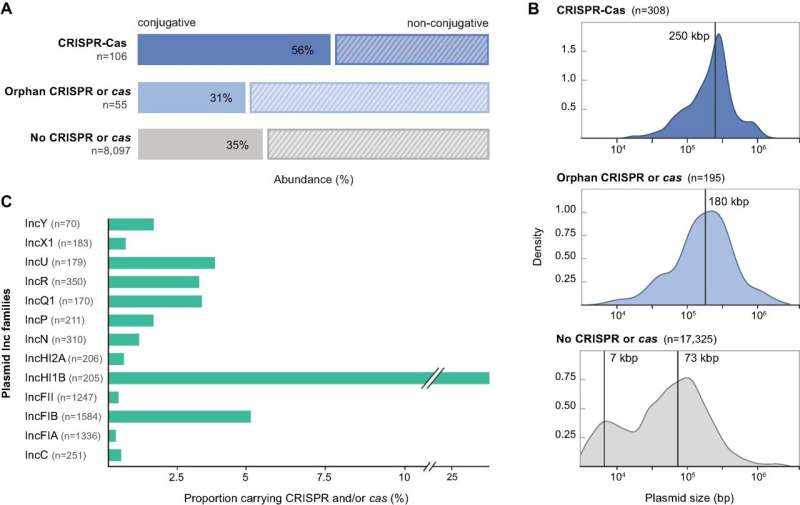
Features of plasmids encoding CRISPR-Cas components. (A) Mobility predictions for the collection of non-redundant proteobacterial plasmids analyzed in this study, presented according to their CRISPR-Cas contents: complete CRISPR-Cas loci, orphan CRISPRs or cas, and no CRISPR or cas. (B) Size distributions for the collection of plasmid genomes carrying complete CRISPR-Cas loci, orphan arrays, solo cas operons and no CRISPR or cas genes. Vertical lines indicate median plasmid size for the unimodal distributions and estimated means from a 2-component gaussian mixture model for the bimodal distribution. Densities are computed with default parameters in base R. (C) Distribution of plasmid incompatibility groups within the Inc-typeable fraction of the complete plasmid dataset and relative abundance of the subset encoding CRISPR-Cas loci. Single plasmids can belong to more than one Inc group. Only Inc groups containing more than 10 plasmids are shown. Credit: Nucleic Acids Research DOI: 10.1093/nar/gkab859
CRISPR-Cas has become somewhat of a superstar over the past decade as a gene editing tool with revolutionary potential, especially in the health sciences. Originally known as an immune defense in bacteria, naturally occurring CRISPR-Cas has proven more diverse and versatile in nature than scientific researchers once believed. Now, a group of researchers at the University of Copenhagen's Department of Biology has investigated the prevalence of CRISPR-Cas systems in plasmids.
The researchers examined more than 30,000 complete plasmid genomes and found CRISPR-Cas in about three percent of them—a high proportion even when compared to bacteria. They also found various representatives of as many as five out of the six known types of CRISPR-Cas in the plasmid genomes studied. The results demonstrate that CRISPR-Cas systems are both widespread and diverse in plasmids and, interestingly, that the vast majority of them target other plasmids.
"In part, this is exciting because it supports a more recent understanding of plasmids as having a higher degree of autonomy from their host cells, typically bacteria. But also, because in the long run, it can open up pathways to combat virulence and resistance in bacteria, which plasmids help spread," explains Rafael Pinilla-Redondo, who is one of the study's main researchers and based at the University of Copenhagen's Department of Biology.
CRISPR acts like a genomic GPS, where a stored memory of foreign DNA fragments can be used to locate a target for Cas proteins, the "genetic scissors". In the majority of the study's findings, it was DNA from other plasmids that was discovered in the immune memory of the CRISPR-Cas systems—i.e. placed in the crosshairs.
Part of a paradigm shift
According to the researchers, this suggests a struggle for resources among plasmids, where plasmids serve their own interests by working actively to prevent other plasmids from accessing the host bacterium in which they reside. In this battle, they use CRISPR as a weapon.
The researchers had the opportunity to simultaneously examine the host bacteria of the 30,000+ plasmid dataset for the same CRISPR-Cas sequences. The idea was to study whether the sequences found in plasmids mirrored the CRISPR contents in host cells, but this was generally not the case.
"Our findings suggest that plasmids have a high degree of autonomy from the bacteria in which they live. While plasmids depend on their host, they are also genetically independent entities that serve their own interests. Their different CRISPR-Cas content is a great example of this autonomy," says Rafael Pinilla-Redondo.
The new research results will contribute to what the researchers consider a paradigm shift in microbiology. In microbiology, gene flow or gene transfer refers to when genetic material moves between cells, mediated mobile genetic elements. While some mobile genetic elements are allowed to slip in, to the benefit of a cell, others are stopped because they are detrimental. The common understanding has long been that bacteria control gene flow.
The paradigm shift points to an understanding where bacteria actually play a far less important role influencing gene flow.
"What was once presumed to be bacteria fighting to protect themselves from genetic parasites, like viruses and plasmids, is far more complex. Perhaps, it should be better understood as the parasites fighting among themselves, for example, over which ones should be allowed to live behind a cow's ear," explains Rafael Pinilla-Redondo.
The possibility for new weapons against antibiotic resistance
The new knowledge about how plasmids use CRISPR may impact how we combat dangerous bacteria in the future. Plasmids are key to the spread of harmful genes between bacteria through what is known as horizontal gene transfer.
The proliferation of genetic material is crucial for bacteria's ability to adapt to new environments and challenges. From an antibiotic resistant bacterium, a plasmid can copy itself and transfer that property to surrounding bacteria as part of its own DNA.
As such, battles between plasmids can help researchers learn more about how to fight them too.
"By understanding how plasmids compete among themselves, we may be able to learn how to slow them down and thereby slow the spread of antibiotic resistance and virulent, harmful properties between bacteria," says Søren Johannes Sørensen, a microbiology professor and co-author of the research article.
"In the long term, it's possible that we will be able to make the strategies of plasmids our own and use them as tools. Without borrowing from nature, we would be quite limited. But if we can learn about the strengths and weaknesses of the plasmids from themselves, opportunities will arise," he says.
What does CRISPR-Cas mean?
DNA fragments (CRISPR) and Cas protein scissors (e.g., Cas9) can locate specific DNA sequences and cut them.
It is predicted that CRISPR-Cas will play a revolutionary role as a gene editing tool, not least in the health sciences, for the treatment of genetic disorders, among other things.
CRISPR systems were originally seen as an immune system for bacteria, particularly against viruses. However, many researchers now see CRISPR-Cas as a "Guns for Hire" tool that can be deployed for a multitude of purposes, by many different actors, including bacteria, plasmids—and humans.
What is a plasmid?
A plasmid is a small ring-shaped DNA molecule, a so-called mobile genetic element, found in bacteria and certain other types of microorganisms.
It is reminiscent of viruses, since both are parasites within cells. Plasmids can reproduce independently of the host cell and often provide benefits to the host cell.
Among other things, they can give or transfer genetic properties to a bacterium, such as by making it antibiotic-resistant or pathogenic, in a process known as horizontal gene transfer.
Plasmids have long been an important tool in molecular biology for, among other things, gene cloning and the introduction of genetic material into bacterial cells.
What is horizontal gene transfer?
Horizontal gene transfer is when one organism transfers genes to another organism that is not its own offspring.
To a great extent, the ability of bacteria to adapt to new environments and challenges depends on the supply of new genes in this way.
The phenomenon is responsible for the looming antibiotic resistance crisis, as bacteria rapidly develop resistance to antibiotics by acquiring resistant genes. It is very often mediated by plasmids, making plasmid proliferation a global public health problem.
About the study: Researchers created a CRISPR scanner
To study the prevalence of CRISPR-Cas in plasmids, the researchers used the largest collection of completely sequenced plasmid genomes, a dataset compiled by researchers around the world.
To manage the hefty quantities of data, the Department of Biology researchers developed software to scan for known CRISPR parts. The program, named CRISPRCasTyper, has since been made freely available to other researchers.
More information: Rafael Pinilla-Redondo et al, CRISPR-Cas systems are widespread accessory elements across bacterial and archaeal plasmids, Nucleic Acids Research (2021). DOI: 10.1093/nar/gkab859
CRISPRCasTyper: crisprcastyper.crispr.dk/#/submit
Journal information: Nucleic Acids Research
Provided by University of Copenhagen