Mysterious genes survive marine organisms' evolutions
When creatures evolve, their genetic blueprints change. But within genetic material, fragments from a species' most ancient ancestors still lurk.
In a recent publication of the journal Genomics, Carnegie Mellon University researcher Saoirse Foley and colleagues found 14 genes that could provide insight into the origins of marine life.
"We found a set of genes whose functions are unknown, and they don't have any known protein domains," Foley said. "They're spread across a set of marine organisms and are very ancient. This was really surprising."
Foley, the first author on the paper, is the senior bioinformatician at Echinobase, a web-based resource that provides access to genomic, expression and functional data from research related to starfish, sea cucumbers, sea urchins and other types of echinoderms.
"Evolutionarily, echinoderms are old and are a good representation of early emerging animals. They provide an uncomplicated view of how a group of animals may emerge," Foley said.
Foley is a part of Veronica Hinman's lab, which studies echinoderms. Starfish and humans, along with other vertebrates, share a number of similarities in their early development, genome organization and gene content.
"Saoirse had started the project thinking she may be able to detect echinoderm-specific genes that might help us understand the attributes of these animals, so it was very surprising to find such ancient, conserved genes and then to see that they had no known functions," said Hinman, the Dr. Frederick A. Schwertz Distinguished Professor of Life Sciences and head of the Biological Sciences Department of Carnegie Mellon. "It will open an exciting new project to try to understand their roles."
The researchers used phylogenomic approaches to search for orthologous genes (genes from different species that are derived from a common ancestral gene) that didn't have a known function among echinoderms. The researchers then used that dataset to identify genes that share an ancestral relationship across other marine animals such as sponges, anemones and sea squirts. Nonmarine animals such as humans, flies or mice had none of the genes.
"What we found was there are no lineage-specific echinoderm genes," Foley said. "The vast number of genes have a relationship with other animals outside of the echinoderm phylum."
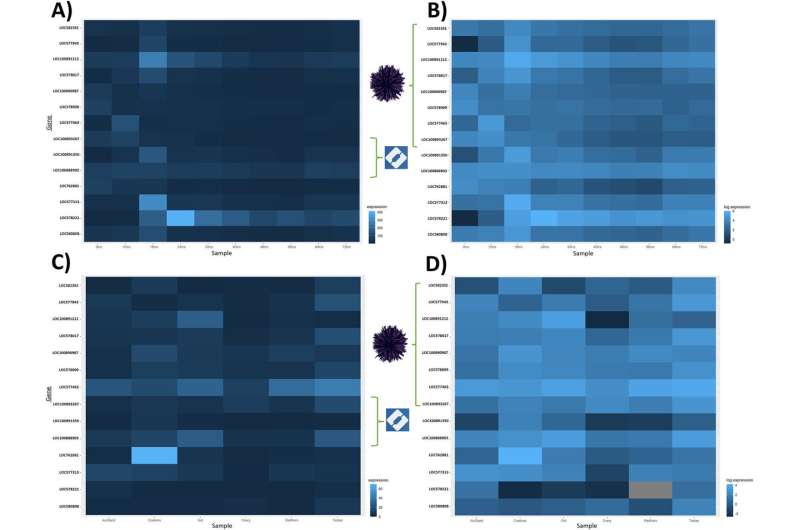
Three echinoderm species were found to have all 14 of the genes: a sea lily (Anneissia japonica), the crown-of-thorns starfish (Acanthaster planci) and purple urchins (Strongylocentrotus purpuratus).
A team from Barcelona—Anna Vlasova, Marina Marcet-Houben and Toni Gabaldón of the Barcelona Supercomputing Center and the Institute for Research in Biomedicine at The Barcelona Institute of Science and Technology—created a family tree using every gene in purple urchins to show how it relates to genes in other species.
"By reconstructing the evolutionary history of every gene in these genomes one can not only find global evolutionary patterns but also identify remarkable exceptions, as we did here," said Gabaldón. "The genes identified are ancient and conserved among marine animals and will be key in understanding how new lineages emerge without the need of acquiring new gene toolkits."
Foley said that while these genes' functions are not currently known, they might be quite important since they all came from a common ancestor. The next step would be to study some of the 14 genes in a laboratory setting to try to discover their functions. Foley said that could involve focusing on purple urchins or another species with some of the genes such as the commonly studied bat star (Patiria miniata).
"The fact that the genes are so old and found only in marine organisms suggests that they are at least performing a marine-specific function," she said. "If they weren't, they're old enough to have been gotten rid of at this point."
More information: Saoirse Foley et al, Evolutionary analyses of genes in Echinodermata offer insights towards the origin of metazoan phyla, Genomics (2022). DOI: 10.1016/j.ygeno.2022.110431
Provided by Carnegie Mellon University